Sexual reproduction is costly.
For starters, only half of the population can bear offspring so the other half has to work hard to make sure they're included in the future gene pool.
Yet there is a payoff beyond the sex jokes sure to follow that statement. Sexual reproduction allows the mixing of parental genomes to generate potentially beneficial new combinations of gene variants that had not previously coexisted on the same strand of DNA, or to separate beneficial mutations from detrimental ones.
In contrast, bacteria reproduce by asexual reproduction, which is more efficient than sexual reproduction since each individual can reproduce simply by dividing. The downside is when the inevitable accumulation of mutations takes its toll, or changes to the environment make gene combinations less well suited than they had once been. Without an exchange mechanism like that provided by sexual reproduction, the bacteria and their offspring are stuck with the same set of genes, for better or for worse.
A new report describes a process by which bacteria can have the best of both worlds - the first genome-wide view of conjugal HGT in bacteria at the nucleotide level.
Using a bacterial species related to the bacterium that causes tuberculosis, Keith Derbyshire, Todd Gray and colleagues, from the Wadsworth Centre in New York, show that a multitude of DNA fragments are simultaneously transferred from a donor bacterial strain to a recipient strain to create new strains that are genetic blends of the parents. The newly-described process, called Distributive Conjugal Transfer, creates patchwork genomes that are different from either parent and different from any siblings. This generates a degree of genome-wide variation similar to that generated in sexual reproduction.
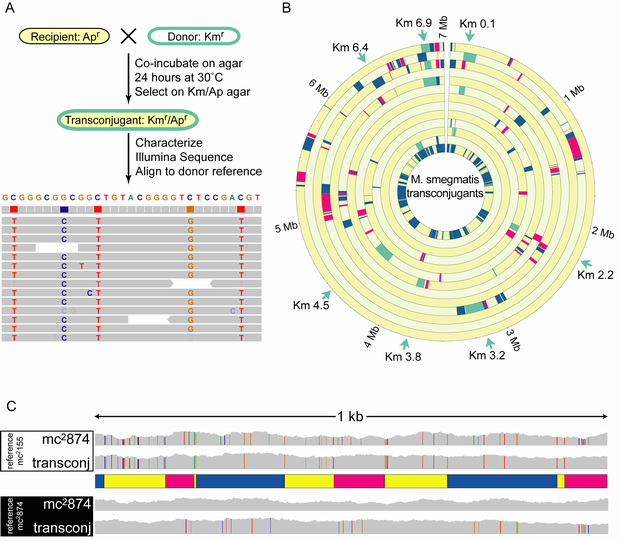
Mycobacterial transconjugant genomes are complex mosaics of their parental strains. (A) Conjugation and genome comparison protocol. Sequence reads for each transconjugant were aligned with the reference donor genome and viewed with IGV. Columns of colored nucleotides mark informative SNPs between the recipient and donor strains, while random colored nucleotides indicate sequence errors. (B) A Circos plot depicts the mosaic nature of 12 M. smegmatis transconjugant genomes. mc2155 donor DNA segments (alternating blue and magenta, or green) replaced homologous recipient sequences (yellow). Positions of integrated kanamycin genes (Km) are shown around the periphery (green arrows), and transferred donor DNA segments containing the Km gene are shown in green. Strain nomenclature is based on the genomic location of the Km gene in Mb, thus Km0.1 is inserted at coordinate 0.1 Mb in mc2155. Strains are from outer to inner circle, respectively: Km6.9e, Km0.1f, Km6.9d, Km3.2, Km6.9c, Km0.1e, Km3.8, Km4.5b, Km2.2a, Km0.1d, Km0.1c, and Km6.4a. The innermost circle is a compilation of all segments of mc2155 DNA, showing that almost all regions of the donor chromosome were transferred despite the small sample size. (C) Microcomplexity of parental SNP profiles at some transconjugant recombination sites. Compiled sequence read landscapes are shown for mc2874 and one transconjugant (Km6.9e) aligned to the mc2155 sequence (top) for a 1 kb segment of the genome (see Table S2, coordinates 470,385–471,385). The presence of informative SNPs (each color represents a different base) indicates recipient sequences, while segments lacking SNPs define donor sequence. Accordingly, parental genotype segments are shown in the schematic below with recipient (yellow) and donor (blue and magenta) genotypes interspersed throughout this 1 kb region. Note that rare cases of isolated recipient SNPs in our designated donor segments are excluded by our stringent criterion requiring two consecutive SNPs to conclusively establish parental origin. The lower panel shows the same sequences aligned to the mc2874 sequence, in which the SNPs now indicate donor sequence. This reciprocal alignment confirms the assignment of donor and recipient sequences in the schematic map.doi:10.1371/journal.pbio.1001602.g001
If the new combination of gene variants make the offspring better suited for growth in its current environment, that bacterium will rapidly divide asexually, out-compete its parental (or sibling) strains and become established as an emerging strain or species in its own right.
Mechanisms by which DNA can be transferred between bacterial genomes have previously been described, but these have been piecemeal, limiting the potential evolutionary benefit, and requiring successive rounds of transfer to create a genome-wide mosaic. However, Distributive Conjugal Transfer generates mosaic genomes overnight. Thus, scientists may have to re-evaluate the projected time it might take for a new mycobacterial strain to evolve, suggest the authors.
The team also exploited a genomic mapping approach similar to those applied in sexually reproducing organisms to localize the mycobacterial genes that determine mating identity. Of the nearly 7,000 genes in the mycobacterial genome, one region spanning just 6 genes appears to be key in determining whether a mycobacterial strain will be a donor or recipient when it comes to mating. This information may help to predict which other bacterial species might participate in this form of gene transfer, and to identify just how widespread the phenomenon is.
The new study suggests that through Distributive Conjugal Transfer, mycobacteria have found a way to reap the benefits of genomic mixing without the dependence and energy costs associated with sexual reproduction. The findings may also shed light on the origin of related mycobacteria that cause tuberculosis. As more mycobacterial genomes are sequenced, a picture is emerging that Distributive Conjugal Transfer may have shuffled the genomes of environmental mycobacteria to create a strain that was particularly well suited for growth in mammalian lungs. However, whether pathogenic mycobacteria, or other non-pathogenic bacteria, actively participate in Distributive Conjugal Transfer has not yet been addressed.
Citation: Gray TA, Krywy JA, Harold J, Palumbo MJ, Derbyshire KM (2013) Distributive Conjugal Transfer in Mycobacteria Generates Progeny with Meiotic-Like Genome-Wide Mosaicism, Allowing Mapping of a Mating Identity Locus. PLoS Biol 11(7): e1001602. doi:10.1371/journal.pbio.1001602





Comments