Molecular biologists and mathematical models frequently don't mix well, especially when the molecular biologists in question were trained before the rise of genomics, back when most labs only needed a computer for designing new vector sequences, writing papers, and checking email. Behind this skepticism is the intuition that biology is extremely messy (true), and difficult to quantify (also true). Also contributing is the long history of the cell as a molecular black box; for a long time, we had no idea what was going on inside the cell in molecular terms (somewhat analogous to doing chemistry without knowing about atoms), and in fact we still don't know the molecular role of a good chunk of the protein-coding genes in the human genome.
Scientists who build quantitative models of molecular biological systems generally face two main criticisms (as I can testify from personal experience, and that of my colleagues), which basically put the would-be model builder in a Catch-22 paradox:
Criticism 1: You can't build a useful mathematical model until you thoroughly understand all of the components of the system, because tomorrow you might discover a new component that will change everything.
Criticism 2: If you thoroughly understand all of the components of the system, then what's the point of your model? It will only add uninteresting detail; you already understand the system well enough.
Both criticisms can be refuted with past examples of success, but molecular biologists are generally unfamiliar with the decades of success stories that prove the usefulness of mathematical models in molecular biology.
One of the great practitioners of quantitative biology was Daniel Koshland, familiar to anyone who has slogged through undergraduate biochemistry as the one who came up with the 'induced fit model' of enzyme-substrate interactions.
Koshland made an argument for quantitative biology in a 1998 essay in Science, "The Era of Pathway Quantification" (subscription required).
Koshland broke down the historical development of molecular biology into three eras:
Knowledge of metabolic processes in cells can be roughly divided into three eras: the Era of Pathway Identification (1890-1950), the Era of Pathway Regulation (1950-1980), and the Era of Pathway Quantification (1980-?). In the first era, the individual steps in the biochemical pathways were identified. In now classical studies, the substrates, products, and enzymes of pathways such as glycolysis,fatty acid metabolism, and nucleic acid metabolism were identified by Emden, Meyerhoff, Warburg, Kornberg, Cori, Brown, Goldstein, and many others (2). In the second era, the control of pathways through feedback, feed-forward, cooperativity, allostery, phosphorylation, and covalent modification was delineated by Pardee, Krebs, Fischer, Stadtman, Jacob, Monod, this author, and many others. In the third era, now in its childhood, the quantification of pathways is being examined to calculate the rates at which metabolites and substrates are produced and degraded in cells and in organs.
What's the benefit of pathway quantification? Koshland uses as his example the phenomenon known as ultrasensitivity in the MAP kinase pathway, which is a molecular pathway for relaying a signal from the outside of a cell (such as the presence of a hormone) down to the inside to generate the appropriate response.
There are a variety of ways that this signal relay system could be rigged up, each producing a different type of response. For example, you could have a system where, as you add more hormone (more signal), you get steadily a increasing cellular response (up until you reach a saturation point). That response would look something like this (except for part where my curve drops down past the origin -what can I say, I'm lazy):
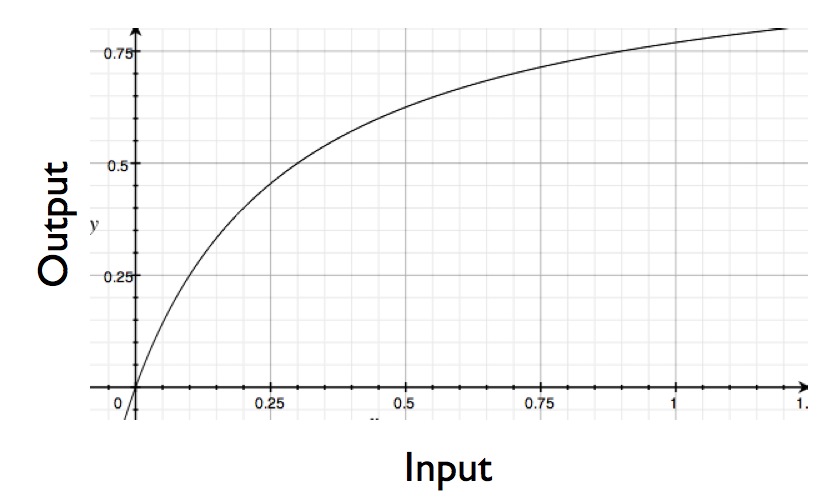
Under a common model of enzyme action (Michaelis-Menten kinetics), Koshland points out that in order to get a 9-fold increase in cellular response, you need a whopping (and biologically rare) 81-fold change in the input signal (such as concentration of your hormone). In other words, a Michaelis-Menten design isn't the best way to rig up your signal sensing system if you want your system to be able to detect changes smaller than an 81-fold increase.
And in fact, we know by experiment that MAP kinase pathways generally aren't so unresponsive. They act more like this:
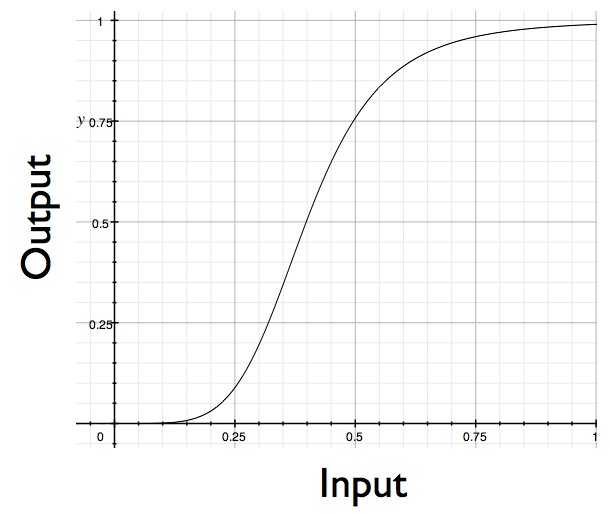
The response curve is sigmoidal, or, to use the jargon, ultrasensitive.
So how do you rig up an ultrasensitive system? Here's the rub: you have no clue unless you bring in the math. There are a variety of ways to generate sigmoidal response curves, which can be understood with a good set of mathematical models. I suppose it's possible to phenomenologically characterize dozens of different ultrasensitive biological systems and attempt to infer some general principles from those experiments. But it's so much easier with a quantitative model - you can make predictions about which system parameters should be important and about the effects of particular mutations. You can, with a little work in Matlab or R, get a good idea of which experiments are likely to be informative.
The work on ultrasensitivity and MAP kinase pathways is a reassuring refutation of the two common criticisms leveled at quantitative biology. A mathematical model, far from being useless if you're missing components of the system, can in fact help you know when you are missing components. A model can tell you when the known components of a system couldn't possibly produce the observed behavior. And your model can explain, in a way that intuitive verbal reasoning can't, how the system manages to produce the observed response.
Koshland was a little optimistic when he suggested that we're in the Era of Pathway Quantification. Genomics and the accompanying high-throughput technologies have put pathway identification and pathway regulation on steroids, and most biologists still make their reputations this way. Pathway quantification is hard, and few biologists can both build the models and do the hard, quantitative experiments to parameterize and test those models. Those who try are faced with the widespread view that pathway quantification is either not feasible or not valuable.
The result is that molecular biologists are too happy being mechanics, as opposed to engineers. A basic mechanic can tell you what the parts of your car do, how, roughly, the force gets conveyed from the pistons to the drive train, and from experience say what kind of an effect a part failure will have on your ability to drive your car. Good mechanics can, with some trial and error experience, juice up your car and make it do something fancy. But most mechanics don't understand a car the way a Mercedes engineer does. They can't build complex computer models of an engine, or come up with the kinds of new designs (for say, fuel efficiency) that an engineer develops.
Biologists need to be more ambitious - we need to understand the cell like an engineer.
In order not to end on a bleak note, check out the work of James Ferrell, at Stanford, for an example of what kind of progress you can find in this field.
Read the feed:





Comments