'Yellow rust' caused by the fungus Puccinia striiformis f. sp. tritici (PST) is one of the plant's major diseases and is found throughout the major wheat-producing areas of the world. Infections lead to significant reductions in both grain quality and yield, with some rare events leading to the loss of an entire crop. New strains of the fungus have recently emerged that adapt to warmer temperatures.
They are more aggressive and have overcome many of the major defensive genes in wheat. Researchers from the John Innes Centre, The Sainsbury Laboratory, TGAC and the National Institute of Agricultural Botany sequenced genetic material from 39 PST-infected samples of wheat collected from 17 UK counties in 2013. By comparing the fungal RNA with fungal genetic information from previously prevalent populations between 1978 and 2011, they showed that there has been a rapid and dramatic shift in the PST population that could have serious implications for wheat production in the UK.
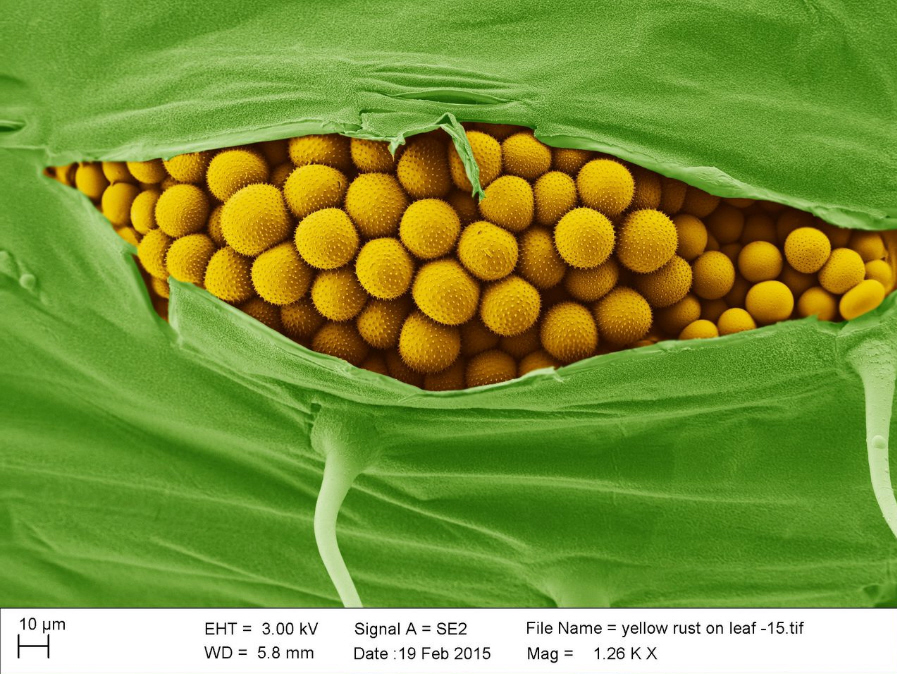
Electron microscopy yellow rust. Credit: Kim Findlay, John Innes Centre
The 2013 PST samples showed more genetic variation and diversity, reflecting an increase in the evolutionary potential in the UK pathogen population that could enhance their ability to overcome disease resistance in wheat.
Of the samples, 11 were also genetically similar to a PST strain called "Warrior". The strain emerged in 2011 as a serious threat to European wheat production due to its virulence on an array of previously resistant wheat varieties. This indicates that a diverse PST population containing the "Warrior" strain is now prevalent across the UK.
This new diagnostic technique, called "field pathogenomics", could be applied internationally to respond to the spread of a wide variety of plant diseases. By rapidly pinpointing a fungus's genetic make-up from field samples, the technique is able to confirm outbreaks on particular wheat varieties and provides an efficient means of confirming whether previously resistant wheat varieties have been broken by virulent strains of the pathogen. This is in contrast to current techniques which can be lengthy, costly and are only able to sample a relatively small proportion of the fungal population.
The data collection and analysis took just a few months to produce from sample collections from the field, demonstrating the potential for the method to reduce delays and transform current disease surveillance systems. The highly detailed information that is generated could help inform disease incidence predictions and agricultural practices.
Citation: Amelia Hubbard, Clare M Lewis, Kentaro Yoshida, Ricardo H Ramirez-Gonzalez, Claude de Vallavieille-Pope, Jane Thomas, Sophien Kamoun, Rosemary Bayles, Cristobal Uauy and Diane G.O. Saunders, Field pathogenomics reveals the emergence of a diverse wheat yellow rust population Genome Biology 2015 DOI: 10.1186/s13059-015-0590-8





Comments