Retroviral proteases are a class of enzymes that play an important role in the maturation and proliferation of the AIDS virus. As such, this class of enzymes is a subject of intense research. The efforts, however, were hindered by a fundamental problem: nobody knew exactly what these enzymes looked like.
To resolve this issue, an online game, Foldit, was created by the University of WashingtonCenter for Game Science, in collaboration with the Baker lab in the Department of Biochemistry of the same university. The game provided direct manipulation tools and the assistance of a program called Rosetta in order to rotate amino acid chains in 3D space and eventually fold a workable protein model. The results were impressive. After three weeks, the structure of the protein was deciphered (see figure 1).
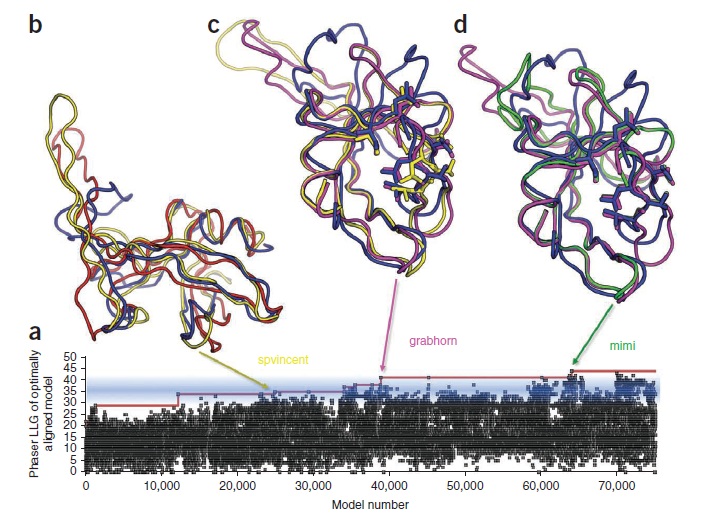
Figure 1: Progress of the gamers in finding the protein's structure. The blue strand in b, c and d indicates the eventual structure. The steps towards this were red to yellow (b), yellow to magenta (c) and magenta to green (d).
(Source: Khatib et al.; 2011)
In the words of the authors:
Remarkably, Foldit players were able to generate models of sufficient quality for successful molecular replacement and subsequent structure determination. The refined structure provides new insights for the design of antiretroviral drugs.
And this might be only the beginning. The (flawed) shape of proteins is an important topic of enquiry in research concerning cancer, Alzheimer’s disease, immune deficiencies and several other disorders. Even research into the development of biofuels relies on knowledge of protein shapes. This success of the online game suggests that online games can provide a powerful tool for scientific discovery. In the words of Seth Cooper, the game’s lead designer:
People have spatial reasoning skills, something computers are not yet good at. Games provide a framework for bringing together the strengths of computers and humans.
The researchers conclude:
The critical role of Foldit players in the solution of the M-PMV PR structure shows the power of online games to channel human intuition and three-dimensional pattern-matching skills to solve challenging scientific problems. Although much attention has recently been given to the potential of crowdsourcing and game playing, this is the first instance that we are aware of in which online gamers solved a longstanding scientific problem. These results indicate the potential for integrating video games into the real-world scientific process: the ingenuity of game players is a formidable force that, if properly directed, can be used to solve a wide range of scientific problems.
References
Khatib, F.; DiMaio, F.; Foldit Contenders Group, Foldit Void Crushers Group, Cooper, S.; Kazmierczyk, M.; Gilski, M.; Krzywda, S.; Zabranska, H.; Pichova, I.; Thompson,J.; Popovic, Z.; Jaskolski, M and Baker, D. (2011). Crystal structure of a monomeric retroviral protease solved by protein folding game players. Nature Structural&Molecular Biology. Published online 18 September 2011. Doi:10.1038/nsmb.2119.
University of Washington, News.





Comments