On top of all of this, there is another code - not so explicitly defined as the genetic code, but an additional layer of information nonetheless. The DNA inside of a eukaryotic cell is tightly packaged, wrapped up around 8-protein cores called histones. This total package, DNA wrapped around a histone octamer, is called a nucleosome. Nucleosomes are sometimes compared to thread wrapped around a spool, but this comparison is way off - DNA isn't nearly as flexible as thread, and that poses a challenge to the cell's genetic regulatory apparatus.
Instead of being like thread, DNA is a little like your typical $1 plastic snake toy - flexible only in certain places (which are determined by the DNA sequence):

Inside the cell, relatively stiff molecules of DNA get wrapped around nucleosomes in an amazing feat of macromolecular origami:
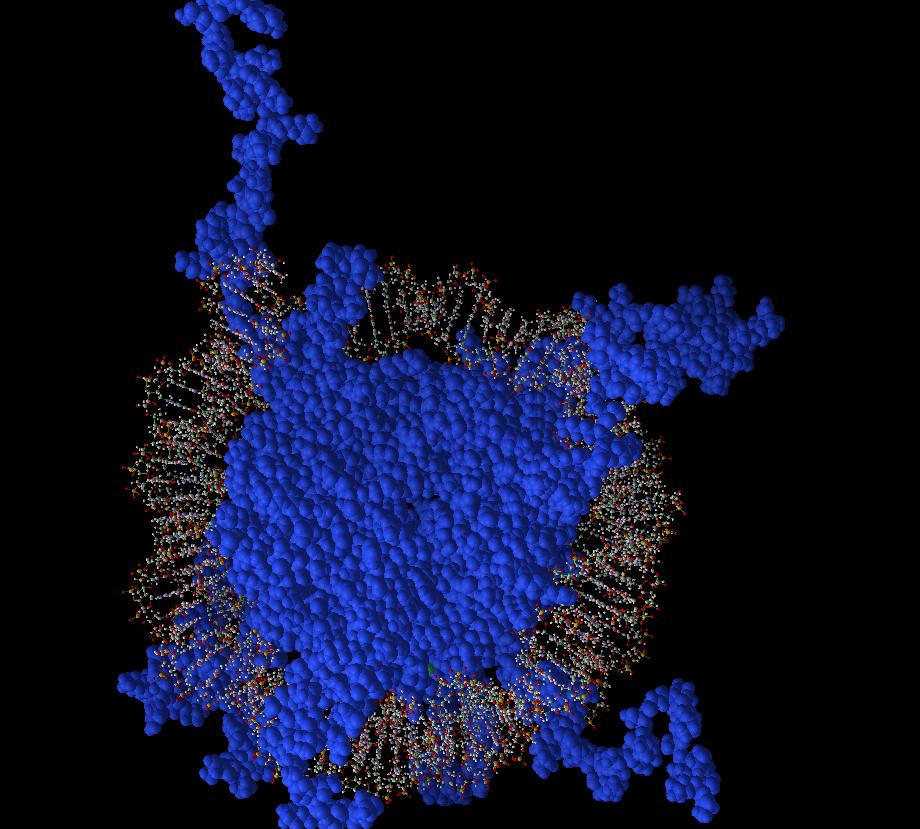
A nucleosome: DNA wrapped around a blue histone core.
This wrapping puts a lot of stress on the DNA. Since some DNA sequences are more flexible than others, nucleosomes tend to occur more frequently at some places in the genome, whereas other stretches of chromosome are nucleosome-poor. Here, then, is another opportunity for a code: the sequence of the DNA influences the placement of nucleosomes.
Nucleosome density matters, because tightly-wrapped DNA is inaccessible to the regulatory proteins that switch on and transcribe genes, and thus tightly-wrapped genes tend to be shut off. The positioning of nucleosomes can clearly have an impact on how genes are expressed, although just how this works isn't clear - which is why nucleosomes have been frequently invoked as a catch-all explanation for anything about gene expression we don't understand.
But just what kind of an impact do nucleosomes have? Has the positioning of nucleosomes been fine-tuned by selection to form an additional, highly specific layer of regulatory code on top of the much better-understood regulatory system of transcription factor proteins that control gene expression? Or are nucleosomes instead obstacles that have to be worked around by regulatory proteins?
This is a familiar type of question in biology: is a particular feature of the living world a highly adaptive, specifically selected function, or is it just a side-effect of something else? Nucleosomes clearly exist for a reason - to package up DNA. But are nucleosomes positioned just so for an adaptive reason as well?
You can think of this question in terms of coding in the DNA. We already know of two highly specific codes: 1) The genetic code, which relates DNA sequence of genes to amino acid sequence in proteins. 2) The gene regulatory code: regulatory proteins bind to very specific stretches of DNA, and switch genes on or off. The expression of a gene is determined, to a large degree, by its surrounding regulatory DNA.
What about nucleosomes? Is this a third layer of code that controls gene expression? This question has provoked some occasionally heated arguments about biologists who study gene regulation.
Yes, there is certainly information in the DNA sequence that encodes the positioning of nucleosomes, information that comes largely from the varying stiffness of DNA. And the positioning of nucleosomes can have an impact on how a gene is expressed. Furthermore, nucleosomes are chemically modified in way that alter the expression of the surrounding genes.
The controversy essentially concerns information flow: Are there DNA sequences that specifically position nucleosomes to carry out certain gene regulatory functions? Or does the information flow from the much more specific transcription factor binding sites, which recruit regulatory proteins that then in turn reposition nucleosomes as needed? The argument has gone back and forth:
A genomic code for nucleosome positioning: "This nucleosome positioning code may facilitate specific chromosome functions including transcription factor binding, transcription initiation, and even remodeling of the nucleosomes themselves."
Nucleosome position signals in genomic DNA: "Our analysis suggests that only a subset of nucleosomes are likely to be positioned by intrinsic sequence signals. This observation is consistent with the available experimental data and is inconsistent with the proposal of a nucleosome positioning code."
...evolution of nucleosome encoded-DNA organization: Our analysis suggests that only a subset of nucleosomes are likely to be positioned by intrinsic sequence signals. This observation is consistent with the available experimental data and is inconsistent with the proposal of a nucleosome positioning code."
Mechanisms that specify Promoter Nucleosome Location and Identity: "Despite the power of these descriptive genome-wide studies as well as work that indicates that these characteristics of promoters play key roles in gene regulation, they leave open the question of how these structures are programmed....The finding in this study and in the previous study that the final resting positions of nucleosomes are strongly influenced by ATP-dependent chromatin remodeling mechanisms argues that that the intrinsic affinity of the octamer for underlying DNA sequences is not determinative for the final positioned state."
One group recently set out to answer this question definitively: they took everything except DNA and histones out of the equation, so that other factors couldn't possibly interfere with the arrangement of histones on the DNA. Taking purified histone proteins from chicken, and genomic DNA from yeast, the researchers reconstituted nucleosomes in a test tube. So where are the nucleosomes now, when they can't be repositioned by remodeling proteins? It turns out that nucleosomes were largely positioned in the same spots they're found inside of a cells. DNA determines nucleosome position - which maybe determines gene regulation.
But... the definitive experiment actually hasn't definitively determined the answer. (I'm exaggerating anyway - the researchers didn't claim that this was the experiment to end all questions.) Aside from some criticism of how this particular experiment was done, the question of information flow is still open: nucleosomes in a test tube were not placed in exactly the same positions in which they're found in the cell; there were small differences. Maybe these small differences in positioning are caused by information flowing from regulatory proteins to the nucleosomes - in fact, we know this happens, because there are large protein machines devoted to shuffling nucleosomes around.
And so the argument goes on. Whatever the answer turns out to be (personally I favor the idea that information flow in gene regulation comes largely from transcription factor binding sites), it's clear that DNA encodes an amazing amount of information (along with plenty of noise). DNA doesn't just code for the amino-acid sequences of the proteins that your cells produce; it also codes for how those genes are regulated under particular environmental conditions. In fact, DNA is so information-rich, that you can take a human chromosome, put it into a mouse, and the information from the human DNA, and not the cellular environment of the mouse, will determine gene regulation on the human chromosome. Tracing information flow in DNA is going to keep researchers busy for years to come.





Comments