Here's an example that deals with a big question that we've all encountered, in science magazines, science news stories, technical papers, and talks: what makes us human? Put more biologically, a fascinating question that geneticists and genome biologists are focused on is what are the genetic differences between humans and chimps, our closest relatives, and how do those genetic differences make us so different?
A recent paper from an excellent geneticist at Duke University, Dr. Gregory Wray, identifies differences in gene expression in human, chimp, and macaque brains. Here is the abstract:
Despite striking differences in cognition and behavior between humans and our closest primate relatives, several studies have found little evidence for adaptive change in protein-coding regions of genes expressed primarily in the brain. Instead, changes in gene expression may underlie many cognitive and behavioral differences. Here, we used digital gene expression: tag profiling (here called Tag-Seq, also called DGE:tag profiling) to assess changes in global transcript abundance in the frontal cortex of the brains of 3 humans, 3 chimpanzees, and 3 rhesus macaques. A substantial fraction of transcripts we identified as differentially transcribed among species were not assayed in previous studies based on microarrays. Differentially expressed tags within coding regions are enriched for gene functions involved in synaptic transmission, transport, oxidative phosphorylation, and lipid metabolism. Importantly, because Tag-Seq technology provides strand-specific information about all polyadenlyated transcripts, we were able to assay expression in noncoding intragenic regions, including both sense and antisense noncoding transcripts (relative to nearby genes). We find that many noncoding transcripts are conserved in both location and expression level between species, suggesting a possible functional role. Lastly, we examined the overlap between differential gene expression and signatures of positive selection within putative promoter regions, a sign that these differences represent adaptations during human evolution. Comparative approaches may provide important insights into genes responsible for differences in cognitive functions between humans and nonhuman primates, as well as highlighting new candidate genes for studies investigating neurological disorders.
I've highlighted the usual result in studies like this: you find a list of genes that show rapid evolution, are differentially expressed, or somehow or other pop up in your survey/genetic screen. Most biomedical scientists are familiar with this kind of work: research that ends in a list of genes associated with some trait or process. I've done this kind of work myself. It's an essential first step.
But what can you do with this list of genes? How do molecular differences in synaptic transmission, transport, oxidative phosphorylation, and lipid metabolism make us human and not chimp?
Obviously, this is a hard question, and it can't be answered on the same sweeping scale as the genomic studies that generate these lists of genes in the first place. Getting at the genes involved in a process is a critical first step; however, my dissatisfaction is rooted in the fact that I almost never encounter research that gets us back to the original traits that motivated the study.
I was struck by this phenomenon when listening to a very nice, classic genetics talk in our weekly departmental seminar the other week. Dr. William Pavan, a mouse geneticist at the NIH, studies melanocytes, the pigmentation cells that produce patterns of skin and hair color, and which go awry in melanoma and a variety of other skin diseases. Dr. Pavan is interested in finding the genes that control the development of melanocytes from a particular population of embryonic stem cells.
To find these melanocyte-regulatory genes, Dr. Pavan generates mutant mice that have pigmentation defects: they go gray early, they develop a white patch of fur on the forehead, or the abdomen, or some other part of their coats. These pigmentation defects are a phenotypic sign that something is going wrong with the melanocytes in these mice. Interestingly, the pigmentation defects in these mutant mice are incredibly precise: a particular mutation puts a white patch of fur in the same place on every mouse harboring the mutation; these are not random patches of white fur.
So Dr. Pavan has a bunch of mutant mice with pigmentation phenotypes. Once you've got a mutant mouse with a reproducible pigmentation defect, the next step is to identify the gene that carries the mutation. Geneticists are very good at gene finding: given an abnormal trait, they can, in many cases, track down the particular genetic alteration that produces that trait.
In other words, we're good finding causal links between traits and genes. And thanks to genomics, we know the molecular function of many of those genes. Once a geneticist finds a gene linked to a trait, it's often possible to look up the molecular function of that gene in a database. At that point, the ball is picked up by the molecular biologists and biochemists, who've got the tools to work out, in great detail, the molecular function of the products encoded by specific genes. This generates a new causal link: we go from genetics to molecular biology. We can then explain the molecular effects: mutating a gene at a particular point results in a protein that does x.
This is where things tend to dead-end: on the smallest scale. The chain of explanation is never brought back to the initial trait that set off the research. Unfortunately, it's extremely difficult to go from detailed molecular biology to a physical explanation of the phenotype: You have a mouse with a precisely positioned white forehead patch. That white forehead patch is somehow caused by a mutation in gene X. Gene X, as a glance at a genomic database will tell you, is an RNA binding protein. A biochemist figures out that the white-patch-causing mutation reduces the affinity of this RNA binding protein for RNA.
After all of this, we still don't actually understand why a mutation in an RNA-binding protein causes a white patch only in the forehead, because we fail to carry our explanations from molecular biology to macroscopic traits:
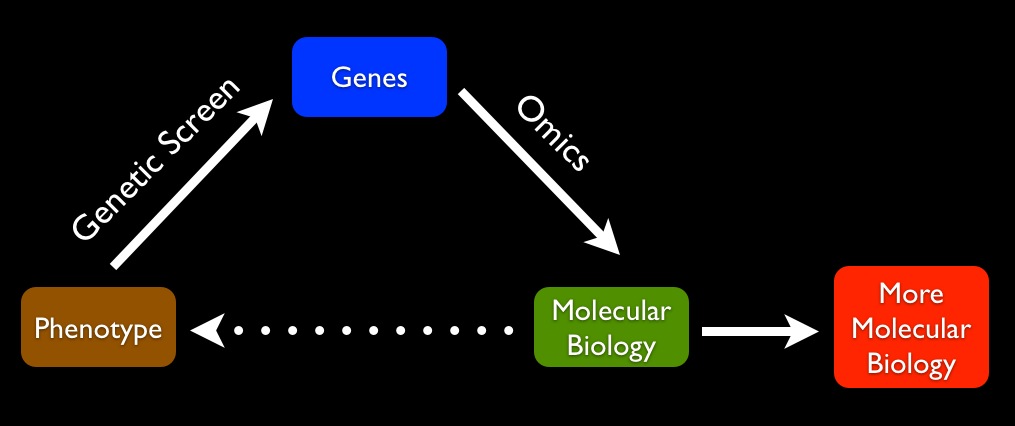
This explanatory gap leaves me dissatisfied after most genetics and genomics talks I hear. Unlike physicists, who in many cases are very good at explaining the properties of solids, liquids, and gases in terms of the properties of atoms, biologists generally (with some exceptions in simple cases) cannot account for macroscopic traits in terms of the physical properties of the molecules of the cell. After typical genetic/genomic studies, you're left with a list of genes that somehow, through a set of physical interactions, produce a phenotype, but just how that works is never explained. Almost all explanation is focused on going from large to ever smaller scales. We almost never get from molecular biology to phenotype. In other words, we still really don't understand how a developing, living, environment-sensing organism arises from a bunch of biophysical interactions.
Read the feed:





Comments