Recently, a lot of attention has gone to epigenetics, or heritable changes in gene expression caused by mechanisms other than changes in DNA sequence. An example of such a mechanism is DNA methylation, where a methyl group (CH3) binds to cytosine or adenine. Epigenetic changes might explain the discrepancies that are being discovered between what the genetic code dictates and how an organism actually looks and functions, ranging from flower shape to the tendency for obesity.
Now, a new study in Science has taken a look at the changes in the epigenetic code over several generations. To do this, the researchers used Arabidopsis thaliana, one of the most well-studied plants in the world. The plants that were used were all clones from a single ancestor, so their DNA sequences were essentially identical (see figure 1).
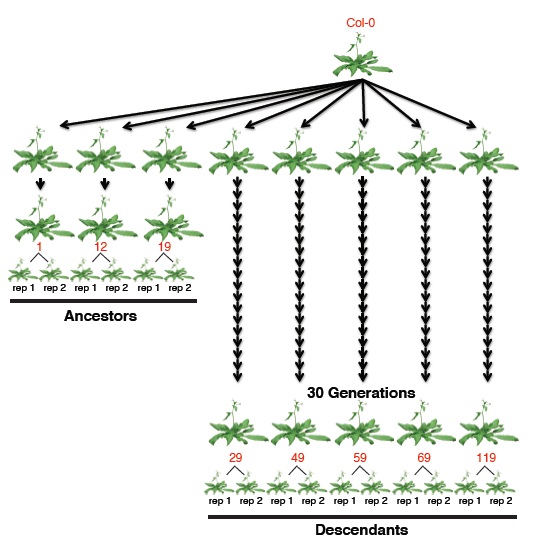
Figure 1: Scheme of the study's design. A single founder line was used. Three replicate ancestral lines are separated from the original by three generations, five replicate descendant lines are separated from the original founder by thirty generations.
(Source: Schmitz et al., 2011)
Why use clones? In the words of Professor Joseph Ecker, who led the research team:
You couldn't do this kind of study in humans, because our DNA gets shuffled each generation. Unlike people, some plants are easily cloned, so we can see the epigenetic signature without all the genetic noise.
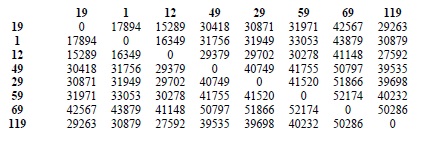
Over the next 30 generations, the researchers from the Salk Institute for Biological Studies and the Scripps Research Institute mapped the epigenome and investigated how this had changed. It turns out that as many as a few thousand methylation sites were altered each generation (see table 1). While still a rather small proportion of the total number of potential methylation sites, this rate is far higher than that of spontaneous changes in the DNA sequence. This might indicate that the epigenetic code is a lot more 'flexible' than the genetic one.
Table 1: Number of epigenetic differences between lines. A difference occurs when one sample has a methylated CG at a particular position and the other sample an unmethylated one. Only positions that agreed within replicates and that were covered in all samples were used.
(Source: Schmitz et al., 2011)
Some of these epigenetic changes even turned genes on or off, meaning that the epigenetic code was not only changing rapidly, but also had a noticeable influence on the plants’ form and function.
Further research is planned to elucidate the biochemical mechanisms that lie at the root of these changes and allow them to be passed on to the next generation. The interplay between environment and epigenetic change is also a topic of further inquiry.
The authors conclude:
Thus, transgenerational epigenetic variation in DNA methylation may generate new allelic states that alter transcription, providing a mechanism for phenotypic diversity in the absence of genetic mutation.
References
Salk Institute for Biological Studies, News
Schmitz, R.J.; Schultz, M.D.; Lewsey, M.G.; O’Malley, R.C.; Urich, M.A.; Libiger, O.; Schork, N.J. and Ecker, J.R. (2011). Transgenerational Epigenetic Instability Is a Source of Novel Methylation Variants. Science. Published online September 15, 2011. doi:10.1126/science.1212959.




Comments