University of California - Davis biochemists have shown for the first time that the ergodic theorem can be demonstrated by a collection of individual protein molecules; specifically, a protein that unwinds DNA.
The ergodic theorem, proposed by mathematician George Birkhoff in 1931, holds that if you follow an individual particle over an infinite amount of time, it will go through all the states that are seen in an infinite population at an instant in time. It's a fundamental assumption in statistical mechanics -- but difficult to prove in an experiment.
Using technology invented at UC Davis for watching single enzymes at work, Bian Liu, a graduate student in the Biophysics Graduate Group and professor Steve Kowalczykowski, Department of Microbiology and Molecular Genetics and UC Davis Cancer Center, found that when they paused and restarted a single molecule of the DNA-unwinding enzyme RecBCD, it could restart at any speed achieved by the whole population of enzymes.
Liu and Kowalczykowski weren't attempting to test laws of physics when they began the work. They wanted to know why RecBCD, an enzyme that unwinds DNA in E. coli bacteria, showed so much variability in its rate of action.
RecBCD attaches to and moves along DNA, unwinding the double helix into two separate strands. It has two jobs in the cell: to allow damaged DNA to be repaired, and to break down invading "foreign" DNA from viruses.
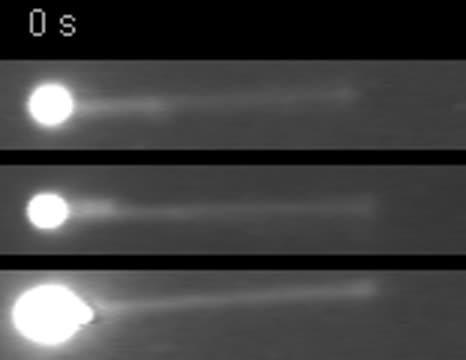
RecBCD enzymes are unwinding DNA at different speeds. The bright ball at left is a bead, the bright strand is a stretch of DNA that shortens as it is unwound by the enzyme. The enzymes show ergodic behavior, supporting an important theory in statistical physics. Credit: Bian Liu, UC Davis
In 2001, Kowalczykowski's laboratory, with the late professor Ronald Baskin at UC Davis, developed a technique to trap single molecules of RecBCD and watch them at work on a strand of DNA in real time. They have since exploited the method to study how DNA is repaired -- in humans, a vital process in protecting against cancer and developmental defects.
"Ever since the original experiments, we've noticed RecBCD molecules have quite a broad range of speeds," Kowalczykowski said.
Liu used the single-molecule visualization technique to measure the rates of hundreds of RecBCD molecules, finding bell-shaped curves for the whole population.
One explanation could be that a large proportion of the proteins were not folded properly and were "trapped" in an inefficient state. However, mild heat or unfolding treatments, which should have allowed the proteins to relax into their correct folded state, had no effect.
RecBCD usually runs for about a minute before stopping spontaneously. Liu found that he could stop the enzyme early by taking away ATP, the chemical fuel that makes the enzyme work.
When he brought back the fuel, he found that the enzymes started up again -- but at a random speed, not related to their previous rate. Overall, the individual RecBCD proteins could restart at any speed within the bell-shaped spread shown by all the proteins.
The experiment shows that RecBCD can move through a wide range of slightly different conformations in which it works at slightly different speeds. However, when it is attached to a step on the DNA ladder, it is locked in shape. Because the time for the enzyme to move from step to step along DNA is shorter than the time it needs to change conformation (about one second), it remains in the same conformation as long as it is moving along DNA, Kowalczykowski said.
What is the point? Why not just have all the enzymes work at one, optimal rate? Having this important enzyme able to operate at a range of speeds might give the cell flexibility to respond to rapidly changing conditions, Kowalczykowski said. For example, degradation of foreign DNA is a process that needs to go quite fast: copying and repairing DNA might require the enzyme to work more slowly, in combination with other proteins.
Citation: Bian Liu, Ronald J. Baskin and Stephen C. Kowalczykowski, 'DNA unwinding heterogeneity by RecBCD results from static molecules able to equilibrate', Nature doi:10.1038/nature12333





Comments