This selective tagging mechanism in natural antibodies has been valuable in engineering antibody-based probes that let them purify and study different types of proteins within cells. One technique, epitope tagging, involves fusing an epitope to a protein of interest and using fluorescently labeled antibodies to make those proteins visible - but only in fixed, dead cells.
Now epitope tagging is getting help - a genetically encoded probe that works in living cells. The researchers call it a "Frankenbody", obviously tweaking the culture wars where regressive environmental activists and academics in the New York University Arthur L. Carter Journalism Institute label everything to do with genetics "Franken-" (not realizing Frankenstein was a graft jump-started by random energy bursts, ancient science they actually accept), because they have taken the binding regions of a normal antibody, the "sticky parts," and grafted them to a different scaffold that remains stable in live cells but retains the specificity of the antibody.
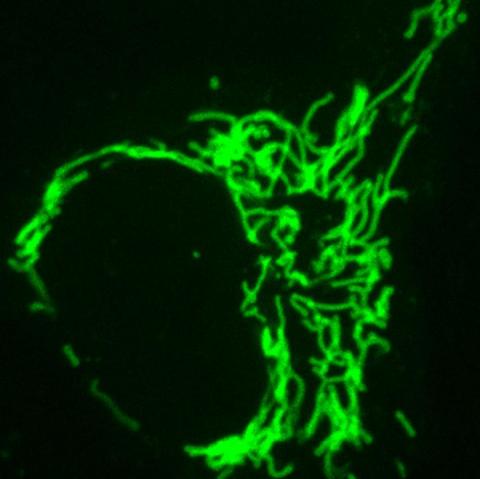
Frankenbody labeling mitochondria. Image: Ning Zhao/Colorado State University
"We're interested in intracellular antibodies because you can use them as imaging reagents in a live cell," said Colorado State University Assistant Professor Tim Stasevich. "You don't need a tag, like a Green Fluorescent Protein, because instead you have this fluorescent antibody that will bind to your protein that you want to visualize."
The new probe would be a useful complement to the green fluorescent protein (GFP), a widespread biochemistry tool and subject of a Nobel Prize that involves genetically fusing a light-up green tag to a protein of interest. However, the GFP is limited by its relatively large size and the time it takes to fluoresce; with the new probe, the tag is smaller and becomes fluorescent faster, so the "birth" of a protein of interest can be captured in real time.
With the goal of making their tool immediately useful, the scientists designed their probe to work with the classic HA tag. HA is a widely used small linear epitope tag that's derived from a portion of the human influenza virus protein hemagglutinin. Now they can image the dynamics of those proteins in live cells. The HA tag is tiny - a chain of just nine amino acids - and the probe is genetically encoded on a plasmid that can be easily transferred into a cell. This is in contrast to traditional antibodies, which can cost a lab several hundreds of dollars per order, suffer from lot-to-lot variability, and are difficult to get into cells. It could be a low-cost solution for protein and RNA translation imaging, with applications including single-protein tracking, single-RNA translation imaging, and amplified fluorescence imaging in zebrafish embryos. All of these experiments are more challenging when using traditional fluorescent protein tags.






Comments