The genome of the medieval form of Yersinia pestis, the pathogen responsible for the Black Death in Europe between 1347 – 1351, has been reconstructed. A surprising finding is that the genes affecting its virulence apparently haven’t changed all that much between then and now.
A research team has used teeth gathered from a burial ground know to contain a lot of Black Death victims to reconstruct the genome of the medieval form of Y. pestis. After the genomic reconstruction, the researchers compared their ‘ancient pathogen’ to the extant forms of the bacterium (see figure 1). In their words:
To place our ancient genome in a phylogenetic context, we characterized all 1,694 previously identified phylogenetically informative positions, and compared those from our ancient organism against aggregate base call data for 17 publicly available Y. pestis genomes and the ancestral Y. pseudotuberculosis.
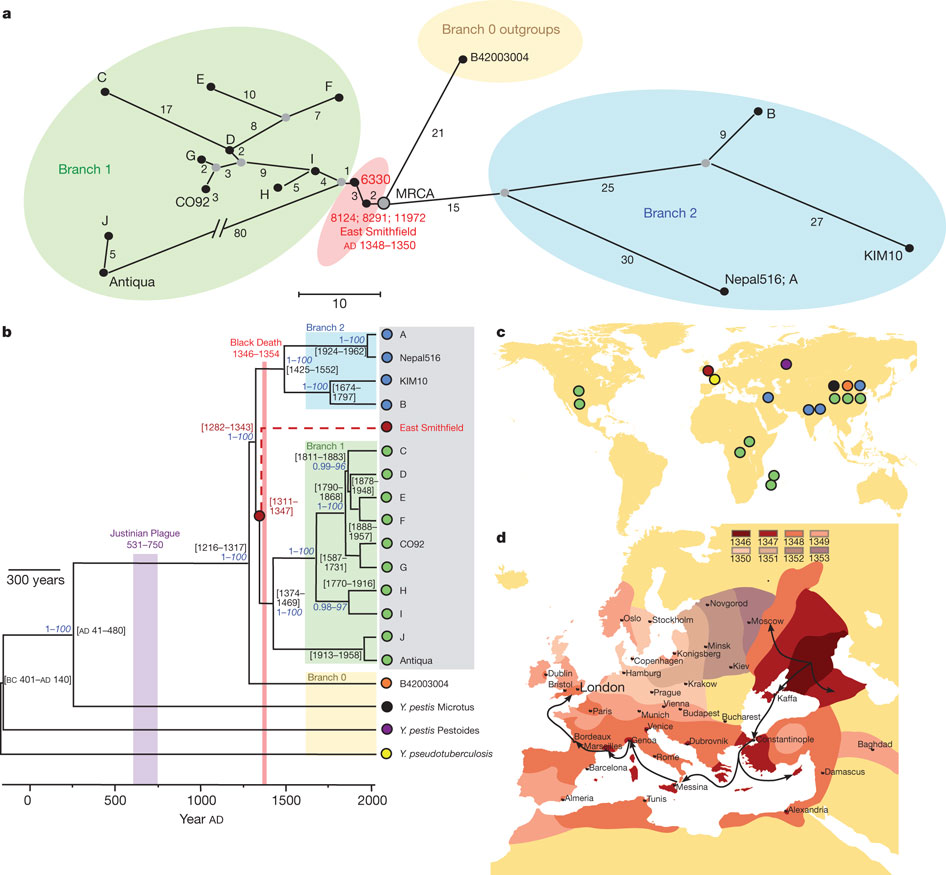
Figure 1: (a) network of ancient and modern forms of the bacterium, (b) phylogenetic tree (grey box indicates known human pathogenic strains), (c) geographical origins of genome sequences used in (a) and (b), (d) spreading of the Black Death.
(Source: Bos et al., 2011)
The thus constructed phylogenetic trees place the reconstructed genome close to the ancestral node of all human strains of the pathogen, which is likely to have arisen between 1282 and 1343, suggesting that the medieval plague was the main introduction of this pathogen into human populations. This, by the way, questions the idea that the Plague of Justinian, from the sixth to eighth century was caused by the same bacterium, as the authors note:
…our temporal estimates imply thatthe pandemic was either caused by a Y. pestis variant that is distinct from all currently circulating strains commonly associated with human infections, or it was another disease altogether.
Furthermore, the genome of Y. pestis, when comparing the reconstructed form with the extant variants, doesn’t seem to have changed all that much, as is mentioned in the authors’ conclusion:
Regardless, although no extant Y. pestis strain possesses the same genetic profile as our ancient organism, our data suggest that few changes in known virulence-associated genes have accrued in the organism’s 660 years of evolution as a human pathogen, further suggesting that its perceived increased virulence in history may not be due to novel fixed point mutations detectable via the analytical approach described here. At our current resolution, we posit that molecular changes in pathogens are but one component of a constellation of factors contributing to changing infectious disease prevalence and severity, where genetics of the host population, climate, vector dynamics, social conditions and synergistic interactions with concurrent diseases should be foremost in discussions of population susceptibility to infectious disease and host–pathogen relationships with reference to Y. pestis infections.
Reference
Bos, K.I.; Schuenemann, V.E.; Golding,G.B.; Burbano, H.A.; Waglechner, N.; Coombes, B.K.; McPhee, J.B.; DeWitte,S.N.; Meyer, M.; Schmedes, S.; Wood, J.; Earn, D.J.D.; Herring, D.A.; Bauer,P.; Poinar, H.N. and Krause, J. (2011). A draft genome of Yersinia pestis from victims of the Black Death. Nature. Published online 12 October. Doi:10.1038/nature.10549. (Click here. Kudos to Nature for making it open access.)




Comments