The truncated IRGM gene is one of only two genes of its type remaining in humans. The genes are Immune-Related GTPases, a kind of gene that helps mammals resist germs like tuberculosis and salmonella that try to invade cells. Unlike humans, most other mammals have several genes of this type. Mice, for example, have 21 Immune-Related GTPases. Medical interest in this gene ignited recently, when scientists associated specific IRGM mutations with the risk of Crohn's disease, an inflammatory digestive disorder.
In a new study, researchers from Evan Eichler's genome science laboratory at the University of Washington and the Howard Hughes Medical Institute reconstructed the evolutionary history of the IRGM locus within primates. They found that most of the gene cluster was eliminated by going from multiple copies to a sole copy early in primate evolution, approximately 50 million years ago. Comparisons of Old World and New World monkey species suggest that the remaining copy died in their common ancestor.
The gene remnant continued to be inherited through millions of years of evolution. Then, in the common ancestor of humans and great apes, something unexpected happened. Once again the gene could be read to produce proteins. Evidence suggests that this change coincided with a retrovirus insertion in the ancestral genome.
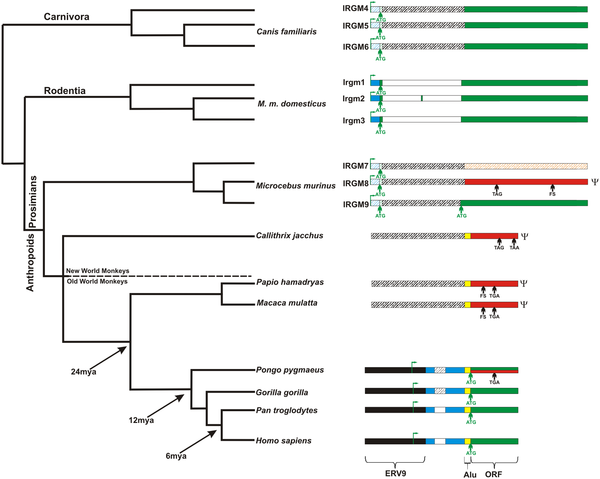
The structures of the IRGM loci are shown in the context of a generally-accepted primate phylogenetic tree. ORF, ERV9, intronic sequence, Alu sequence, and 5′ untranslated region (UTR) depicted in green, black, white, yellow and blue colors respectively. A red color denotes pseudogenes based on the accumulation of deleterious mutations in the ORF. Shaded orange color indicates an atypical GTPase because of mutations leading to the loss of a canonical GTPase binding motif (see Figure S1). The first ATG codon (green arrow) after the Alu repeat sequence is used as putative start codon for the open reading frame of IRGM. The transcription start site is marked with green flag. FS indicates frameshift mutation. TGA and TAA denote the position of stop codons (arrows). The shaded white, blue and green colors indicate predicted intron, UTR or exon, respectively. The genomic loci are not drawn to scale with the exception of the full-length sequence of IRGM ORF. From Bekpen C, Marques-Bonet T, Alkan C, Antonacci F, Leogrande MB, et al. (2009) Death and Resurrection of the Human IRGM Gene. PLoS Genet 5(3): e1000403. doi:10.1371/journal.pgen.1000403
"The IRGM gene was dead and later resurrected through a complex series of structural events," Eichler said. "These findings tell us that we shouldn't count a gene out until it is completely deleted."
The structural analysis, he added, also suggests a remarkable functional plasticity in genes that experience a variety of evolutionary pressures over time. Such malleability may be especially useful for genes that help in the fight against new or newly resistant infectious agents.
CITATION: Bekpen C, Marques-Bonet T, Alkan C, Antonacci F, Leogrande MB, et al. (2009) Death and Resurrection of the Human IRGM Gene. PLoS Genet 5(3): e1000403. doi:10.1371/journal.pgen.1000403






Comments