You may not think of oceanography when it comes to cancer research but an unexpected discovery in marine biomedical laboratories at the Scripps Institution of Oceanography has led to information on how a marine organism creates a natural product currently being tested to treat cancer in humans.
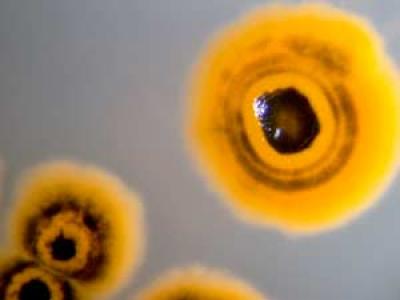
A research team led by Bradley Moore, a professor with the Scripps Oceanography Center for Marine Biotechnology and Biomedicine and Skaggs School of Pharmacy and Pharmaceutical Sciences, and postdoctoral researcher Alessandra Eustáquio, along with their colleagues at the Salk Institute for Biological Studies, discovered an enzyme called SalL inside Salinispora tropica, a promising marine bacterium identified in 1991 by Scripps researchers.
As they describe in the most recent issue of Nature Chemical Biology, the researchers also identified a novel process — a “pathway” — for the way the marine bacterium incorporates a chlorine atom, the key ingredient for triggering its potent cancer-fighting natural product.
Previously known methods for activating chlorine were processed through oxygen-based approaches. The new method, on the other hand, employs a substitution strategy that uses non-oxidized chlorine as it is found in nature, as with common table salt.

“This was a totally unexpected pathway,” said Moore. “There are well over 2,000 chlorinated natural products and this is the first example in which chlorine is assimilated by this kind of pathway,” said Moore.
The Salinispora derivative “salinosporamide A” is currently in phase I human clinical trials for the treatment of multiple myeloma and other cancers. A team led by Moore and Scripps’ Daniel Udwary solved the genome of S. tropica in June, an achievement that helped pave the way for the new discoveries.
Moore believes the discoveries provide a new “road map” for furthering S. tropica’s potential for drug development. Knowing the pathway of how the natural product is made biologically may give biotechnology and pharmaceutical scientists the ability to manipulate key molecules to engineer new versions of Salinispora-derived drugs. Genetic engineering may allow the development of second-generation compounds that can’t be found in nature.
“It’s possible that drug companies could manufacture this type of drug in greater quantities now that we know how nature makes it,” said Moore.
At this point it is unclear how pervasively SalL and its unique biological activation pathway exist in the ocean environment. Chlorine is a major component of seawater, and, according to Moore, a fundamental component of Salinispora’s disease-inhibiting abilities. Salinosporamide A, for example, is 500 times more potent than its chlorine-free analog salinosporamide B.
“The chlorine atom in salinosporamide A is key to the drug’s irreversible binding to its biological target and one of the reasons the drug is so effective against cancer,” said Moore.
According to Eustáquio, finding the enzyme and its new pathway also carries implications for understanding evolutionary developments, including clues for how and why related enzymes are activated in different ways.
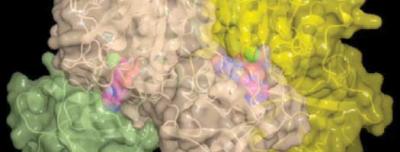
Also joining Moore and Eustáquio in the research were coauthors Florence Pojer and Joseph Noel (of the Howard Hughes Medical Institute, Jack H. Skirball Center for Chemical Biology and Proteomics, The Salk Institute for Biological Studies), who developed high-resolution X-ray structures and other aspects of the research.
The work was supported by the National Oceanic and Atmospheric Administration, the National Institutes of Health and the National Science Foundation.






Comments