Identifying gene ancestry is crucial for computational genomics because genes passed down from a common ancestor tend to perform similar functions in the cell. Scientists exploit this similarity in tasks like predicting gene function, mapping human chromosomal regions to corresponding regions in model organisms, and reconstructing the regulatory circuitry that turns genes on and off.
Although computational biologists have developed methods to identify genes that share a common ancestor, current methods often lead to spurious conclusions when applied genes encode multi-domain proteins. Domains are sequence fragments that encode the basic building blocks of protein structure. Evolution makes new genes by mixing and matching domains in novel combinations, much like a child who builds a house, a car and a helicopter from the same LEGO kit by combining LEGO blocks in different ways.
This process, called domain shuffling, creates complex proteins that perform specific, critical tasks such as cell communication and binding to other cells. When one of these proteins fails, cancer is often the result. Domain shuffling allows rapid evolution of new proteins, but it also makes it close to impossible for scientists to determine their ancestry.
 Opioid Addicts Are Less Likely To Use Legal Opioids At The End Of Their Lives
Opioid Addicts Are Less Likely To Use Legal Opioids At The End Of Their Lives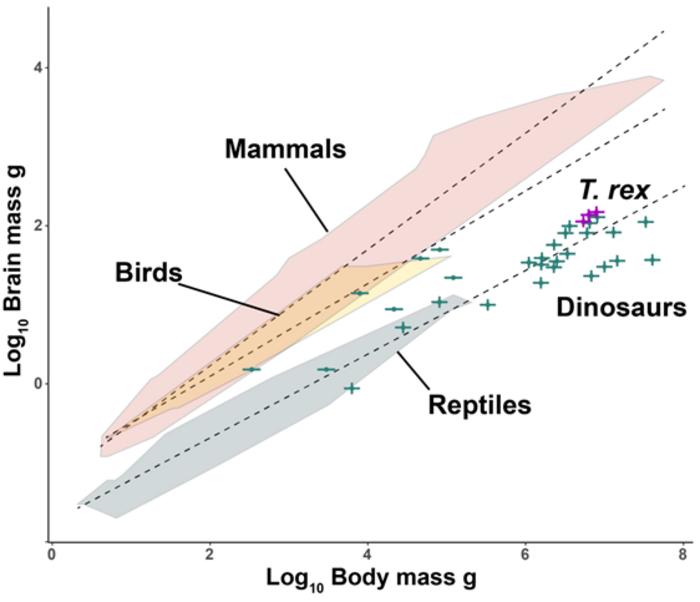 More Like Lizards: Claim That T. Rex Was As Smart As Monkeys Refuted
More Like Lizards: Claim That T. Rex Was As Smart As Monkeys Refuted Study: Caloric Restriction In Humans And Aging
Study: Caloric Restriction In Humans And Aging Science Podcast Or Perish?
Science Podcast Or Perish?