Understanding how and why diversification occurs is important for understanding why there are so many species on Earth. A new study found that similar, or even identical, mutations can occur during diversification in completely separate populations of E. coli evolving over more than 1,000 generations.
The new study involved 3 different populations of bacteria. At the start of the experiment, each population consisted of generalists competing for two different sources of dietary carbon (glucose and acetate), but after 1,200 generations they had evolved into two coexisting types each with a specialized physiology adapted to one of the foods. The researchers were able to sequence the genomes of populations of bacteria frozen at 16 different points during their evolution, and discovered a surprising amount of similarity in their evolution.
"Not only did similar genetic changes occur, but the temporal sequence in which the changes occur over evolutionary time was also similar between the different evolving populations. This 'parallelism' implies that diversification is a deterministic process driven by natural selection," said co-author and University of British Columbia zoologist Prof. Michael Doebeli.
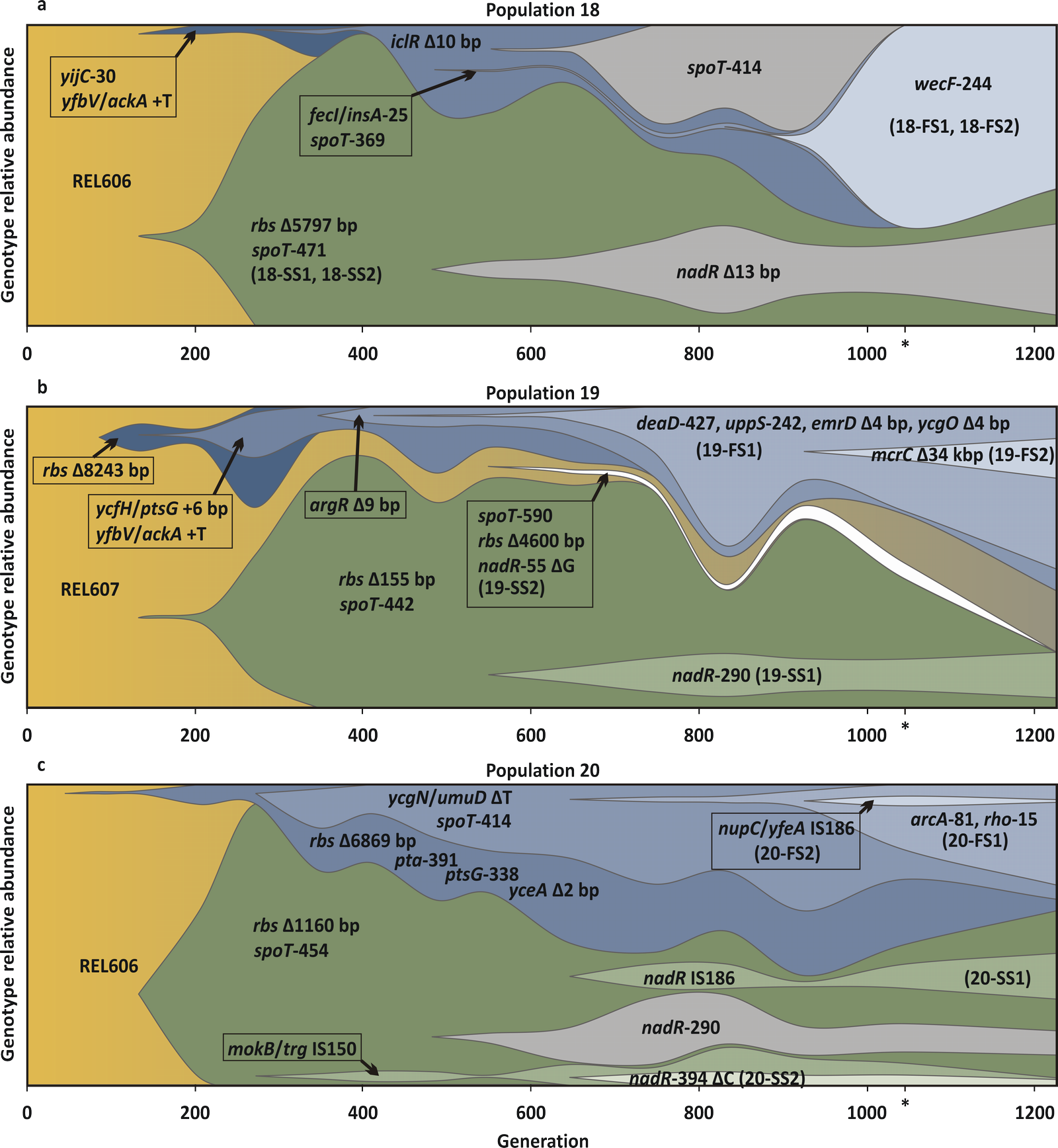
Dynamics of the frequencies of mutations detected in the fossil record of three evolving populations. Shades of blue (above) indicate the mutations associated with FS clones and shades of green (below) indicate the mutations associated with SS clones. Gold indicates ancestral strains (which may include mutations not associated with any sequenced clone). The white region in (b) indicates an independent origin of the SS phenotype. Mutations within a lineage are cumulative—that is, mutations corresponding to lighter regions appear in the genetic background corresponding to the darker regions in which the lighter regions are nested. For example, in population 20 the first mutations to appear in the SS lineage were an 1,160 bp deletion in the rbs operon and a substitution in codon 454 of spoT. An IS150 insertion in the intergenic region between mokB and trg appeared on this background around generation 300 and remained at low frequency for the rest of the experiment. Around generation 650, a single bp deletion in codon 394 of nadR appeared on the rbs Δ1160 bp+spoT-454+mokB/trg IS150 background. Grouping of mutations into lineages was based on their presence together in sequenced clones (in this case 20-SS2), and their order of appearance was inferred from the time point sample in which each was first detected. In addition, three mutations not found in any of the sequenced clones but whose association with SS and FS can be inferred (explained in SI) are shown in gray [FS-associated spoT-414 in (a), and SS-associated nadR-235 in (a) and nadR-290 in (c)]. For visual clarity, mutations of similar frequency within a lineage have been lumped together and their frequencies averaged. Credit: doi:10.1371/journal.pbio.1001490
Recent advances in sequencing technology allowed them to sequence the large numbers of whole bacterial genomes and provide evidence that there is predictability in evolutionary diversity. Any evolutionary process is some combination of predictable and unpredictable processes with random mutations, but seeing the same genetic changes in different populations showed that selection can be deterministic.
"There are about 4.5 million nucleotides in the E. coli genome," said co-author Matthew Herron, research assistant professor at the University of Montana. "Finding in four cases that the exact same change had happened independently in different populations was intriguing."
The researchers argue that a particular form of selection, negative frequency dependence, plays an important role in driving diversification.
When bacteria are either glucose specialists or acetate specialists, a higher density of one type will mean fewer resources for that type, so bacteria specializing on the alternative resource will be at an advantage.
"We think it's likely that some kind of negative frequency dependence—some kind of rare type advantage—is important in many cases of diversification, especially when there's no geographic isolation," Herron said.
As technology advances, they believes that similar experiments in larger organisms will be possible. Some examples of diversification without geographic isolation are known in plants and animals, but it remains to be seen whether or not the underlying evolutionary processes are similar to those in bacteria.
Citation: Herron MD, Doebeli M (2013) Parallel Evolutionary Dynamics of Adaptive Diversification in Escherichia coli. PLoS Biol 11(2): e1001490. doi:10.1371/journal.pbio.1001490






Comments