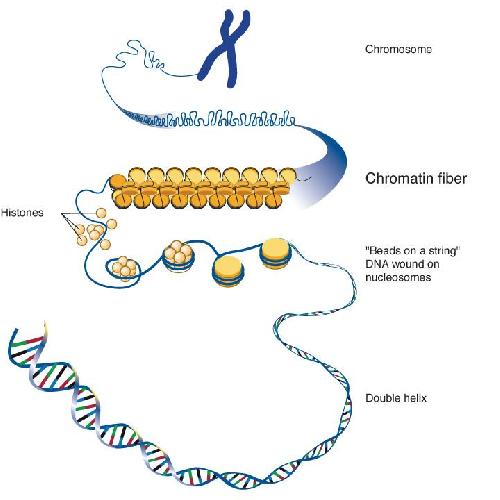
The DNA in our cells is folded into millions of small packets, like beads on a string, allowing two-meter linear DNA genomes to fit into a nucleus of only about 0.01 mm in diameter.
However, these molecular beads, called nucleosomes, render DNA 'unreadable'. They thus need to be temporarily displaced to allow genes to be copied ('transcribed') into the messages that are used to make proteins. How cells ensure appropriate access to 'promoter' DNA, the regions where gene transcription begins, is still poorly understood.
Researchers from the University of Geneva (UNIGE) and the Ecole polytechnique fédérale de Lausanne (EPFL), Switzerland, are studying the mechanisms controlling nucleosome dynamics and how this affects gene expression. They discovered all promoters could be classified into two distinct types differing by their state of nucleosome stability.
One type, characterized by the presence of dynamic, unstable, nucleosomes, is found at highly expressed genes, such as those involved in the control of cell growth and division. The other type, which contains well-known stable nucleosomes, is located at less frequently expressed genes. The interplay of different molecular actors involved in nucleosome destabilization is described in the journal Molecular Cell.
How can about two meters of linear DNA contained in a mammalian cell fit into a nucleus of roughly 0.01 mm diameter? This is done with the help of nucleosomes, basic units made of proteins around which a segment of DNA is wound. When a given gene needs to be transcribed to create new proteins, its promoter region must be unwrapped from the nucleosome so that it can be accessed by the factors involved in initiating the transcription process.
Growth-related genes are highly expressed
"It is important to understand how nucleosomes are moved, ejected or restructured, as this will affect the accessibility of promoter DNA, which in turn influences the expression of the corresponding genes", explains David Shore, professor at the Department of Molecular Biology of the Faculty of Science at UNIGE.
The dynamics of nucleosome formation and positioning in promoters may, for example, help to understand why some genes are highly expressed, such as those involved in normal or malignant growth, while others, such as stress-induced genes, are only rarely expressed under normal conditions.
In collaboration with researchers at the Department of Computer Science (UNIGE) and the Swiss Institute of Bioinformatics at EPFL, David Shore's team has undertaken to characterize nucleosomes present in every gene promoter of yeast DNA.
This unicellular fungus is used as a model organism because it functions in many respects like a mammalian cell. As do human cells, yeast also possesses so-called 'fragile' nucleosomes. Discovered by chance and termed 'fragile' because they don't resist as well as the others to certain enzymes, their nature and function remained elusive.
A unique type of dynamic nucleosome
"We have revealed the existence of two types of promoters, which differ by the presence of 'fragile' nucleosomes," says Slawomir Kubik, first author of the study and researcher at UNIGE.
The scientists also found out that the type of promoter containing 'fragile' nucleosomes is strongly associated with high levels of transcription. They speculate that the dynamic nature of the 'fragile' nucleosome plays an important role in increasing access of the proteins initiating the transcription to the promoter.
"The genome probably stays in a very compact state most of the time, with nucleosomes winding the DNA like a tight spring. We believe that the presence of dynamic nucleosomes at highly expressed genes helps to unwind this spring rapidly and as often as necessary," comments David Shore.
Since many of the genes containing 'fragile' nucleosomes are controlled in a continuous manner by nutrient availability, modulation of promoter nucleosome stability may be a strategy used to coordinate growth-related transcription on a genome-wide level.






Comments