As large, visually-oriented mammals, we have long had a tendency to consider biological diversity primarily in terms of what we can see. There is, however, an entire world of creatures rarely encountered but no less unique and intriguing for it. Sometimes, one only needs the right tools, or the proper motivation, to recognize a group of organisms well worth our attention.
It is in this spirit that I am pleased to introduce you to Hypsibius dujardini , one of about 700 known species in the Phylum Tardigrada , commonly known as a "water bears" due to their ursine appearance.
Literally meaning "slow walkers", tardigrades are tiny water-dwelling animals (typically 0.5-1.0mm) that can be found in droplets of water on mosses or in beach sediments.
Tardigrades are interesting from a physiological perspective because of their extraordinary ability to withstand dessication as metabolically inactive, dried up "tuns". Tardigrades in this state can survive droughts as well as extreme cold, heat, and pressure, and are dispersed by the wind. When once again they come into contact with water, they revive and carry on about their business.
Like most people, I have never encountered a water bear in the wild -- or perhaps I have but did not know it. It was therefore something of a novelty for me to work with some, albeit briefly, about a year ago.
My area of research relates to the question of genome size diversity among animal species. In other words, I am interested in the amounts of DNA carried in the cells of different kinds of animals, a parameter which varies about 7,000-fold among animals. There are, I often point out, some salamanders with 40x more DNA than anyone reading this article.
There are many reasons to study genome size. For example, it represents an important evolutionary question, and has been a subject of active study in this regard for more than half a century. Moreover, the amount of DNA in a species may be related to biological features such as body size, metabolism, and development, and as such it is of interest in a range of disciplines. More pragmatically, the amount of DNA in a genome is a crucial piece of information when one wishes to study a species from a genomic perspective, especially if this will involve sequencing the genome.
Because of their transparency, short generation times, and relationship to other model organisms such as flies and roundworms, tardigrades can be considered good model organisms for studying questions such as how genes regulate development. However, the difficulty in raising these animals in the lab had prevented them from being adopted as model organisms -- that is, until Bob McNuff, who runs the British biological supply company Sciento, determined that Hypsibius dujardini could be reared more or less indefinitely. McNuff's finding is reported together with the work of Bob Goldstein and his student Willow Gabriel, who painstakingly mapped the developmental features of this tiny animal, thereby laying the groundwork for future studies in which the roles of specific regulatory genes can be identified through studying their expression at different stages or by manipulating them and observing the effects relative to untreated specimens.
So promising was Hypsibius dujardini as a model organism that a group of researchers including Goldstein also wished to propose it as a target for complete genome sequencing -- but for that, one needs a genome size estimate. That's where my student Alex Ardila Garcia and I came in.
Using two different methods, flow cytometry and Feulgen image analysis densitometry, we compared the DNA content of nuclei from Hypsibius dujardini to samples from the fly Drosophila melanogaster , and found that the tardigrade's genome is less than half the size of the fly's. In fact, Hypsibius dujardini has the smallest genome I have ever personally analyzed -- a mere 75 million base pairs (for comparison, humans have about 3.2 billion).
The image below shows some Feulgen-stained cells of Hypsibius dujardini , with cells from the fly Drosophila melanogaster in the inset for scale. The lower panel shows a comparison of the same species by flow cytometry analysis.
These results, in combination with the details about tardigrade development have now been published in the journal Developmental Biology
I am also happy to report that Hypsibius dujardini has indeed been listed as a target species for genome sequencing.
Water bears, though part of the largely unseen majority of biological diversity, are now set to reveal major insights into the evolutionary and developmental processes by which that diversity comes to be.
T. Ryan Gregory , William R. Jecka, Corbin D. Jones and Bob Goldstein, The tardigrade Hypsibius dujardini, a new model for studying the evolution of development, Developmental Biology, Volume 312, Issue 2, 15 December 2007, Pages 545-559. doi:10.1016/j.ydbio.2007.09.055 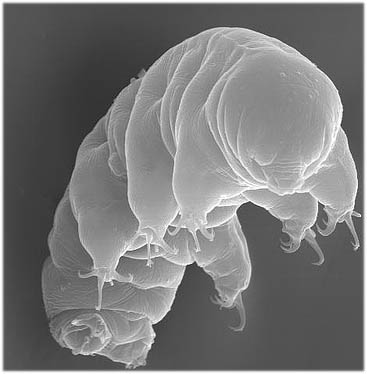







Comments