One of the many aggravations I encounter when reviewing manuscripts is that some authors greatly overstate the applicability of statistically significant patterns they report. For example, a statistically significant pattern in a small comparison of a few animals may be extrapolated in the discussion to the kingdom at large.
Today I was disappointed to see a paper that is soon to come out in Zoology that does the opposite -- i.e. takes a non-significant relationship in a handful of species and pretends that it challenges the importance of broad relationships that have been considered important for decades.
The paper in question is:
Starostova, Z., L. Kratochvil, and M. Flajshans. 2008. Cell size does not always correspond to genome size: phylogenetic analysis in geckos questions optimal DNA theories of genome size evolution. Zoology, in press.
They compared genome size and cell size across 15 geckos and found no correlation. From this, they went on to argue that genome size does not causally influence cell size and that genome size is not under selection due to cell size impacts.
First, let me point out that strong, positive correlations between genome size and cell size have been reported within and across all vertebrate classes including reptiles. So, on a broad scale, the relationship is clear.
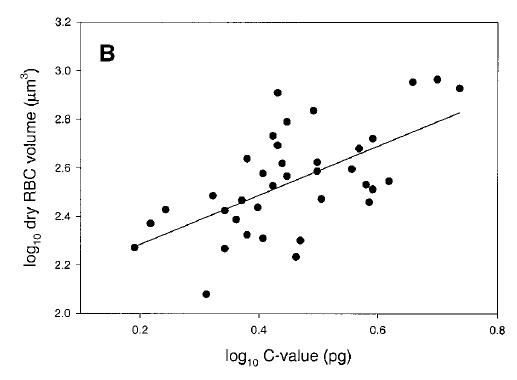
Genome size and cell size in reptiles. From Gregory (2001), based on data from Olmo and Odierna (1982).
Second, let me say that I have issues with their methods. For example, they used DAPI as the fluorochrome, which is base-pair specific and can give biased determinations (they recognize this but assume the species are all the same in AT content). Second, they produced fairly substantial error ranges in their measurements given that these were all raised in the lab or obtained from pet shops and not taken from different wild populations (i.e., the variability between conspecifics is probably artifact). Third, they counted "forms" of the same species from different places as being independent in their analyses -- so it wasn't 15 species, rather it was 12 species with several represented by multiple points.
These are not the main problems, though. The first is that they clearly had outliers in the dataset. In particular, Coleomyx brevis (CB) and Coleomyx variegatus (CV) have "large" (~2pg) genomes but comparatively small cells. I don't think I even need to draw the line through the remaining points, but in case eyeball statistics don't do it, the correlation is highly significant without them (r = 0.74, p < 0.006) (they recognize this, too, but note the title they chose for the paper nonetheless).
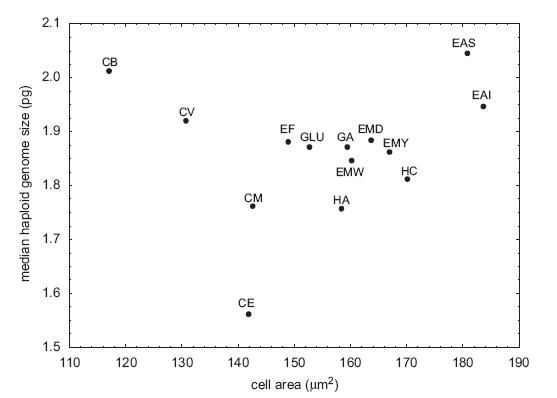
From Starostova et al. (2008).
So, how can this be explained? Well, you have to know something about nucleotypic theory, which these authors actually did mention. It's not genome size all alone that is the determining factor -- nucleus size is critical. The "nucleotype" is defined as "that condition of the nucleus that affects the phenotype independently of the informational content of the DNA” (Bennett 1971). As has been pointed out repeatedly (e.g., by me, Cavalier-Smith, Bennett, and others), the compaction level of DNA in the nucleus adds a second dimension to the relationship. More DNA is one thing, but if it is compressed into a tightly packed, reduced nucleus, then cell size may still be small.
That leads to the second major problem. Looking at the data reported in a previous study (Starostova et al 2005), there is no correlation between genome size and nucleus size. There is, however, a positive correlation between nucleus size and cell size across these reptiles.
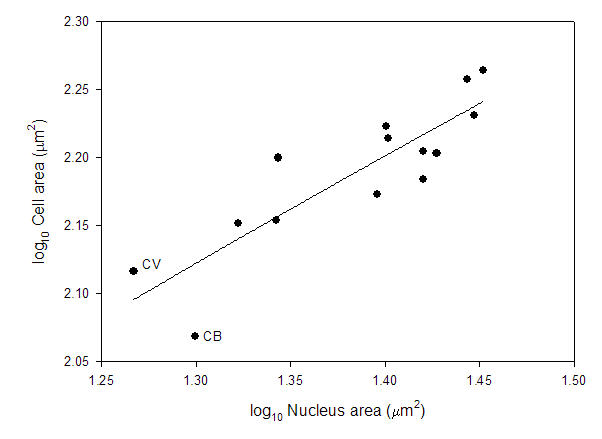
Based on databy Starostova et al. (2005).
The two outliers in the genome size vs cell size comparison have more compact nuclei and this allows smaller cell sizes with larger genome size. Cell size is correlated with body size in these geckos, and these two species are "dwarfs" (~4.5g) relative to other species (as big as ~90g). So, there could very easily be selection for reduced cell size which, in this narrow range in DNA amount, was met by a compaction of the nucleus rather than a loss of DNA.
This actually reinforces the strength of nucleotypic theory.
______
Bennett, M.D. 1971. The duration of meiosis. Proceedings of the Royal Society of London B 178: 277-299.
Gregory, T.R. 2001. The bigger the C-value, the larger the cell: genome size and red blood cell size in vertebrates. Blood Cells, Molecules, and Diseases 27: 830-843.
Olmo, E. and G. Odierna. 1982. Relationships between DNA content and cell morphometric parameters in reptiles. Basic and Applied Histochemistry 26: 27-34.
Starostova, Z., L. Kratchovil, and D. Frynta. 2005. Dwarf and giant geckos from the cellular perspective: the bigger the animal, the bigger its erythrocytes? Functional Ecology 19: 744-749.




Comments