The child first identifies the target: peas. “I do not like peas.” Cautiously spooning through the dark broth, she scans the material in the bowl to determine if peas are present. When the adults aren’t looking, the resourceful child might even poke her fingers into the liquid to assist with pea detection. Characteristics of various components are distinguished by her eyes and fingers and interpreted by her brain for identification. Orange, circular: carrots. White, slimy: noodles. Green, spherical: peas. Peas. “Yuck!!! Peas!!!”
Similarly, scientists would like to scan biological samples like blood, urine, and cell lysates for a variety of components, called biomarkers, which are indicators of cancer and other diseases. However these indicators—proteins, RNA, hormones, and other small molecules—are, at most, one billionth of one billionth the size of the average pea that a child might skillfully identify in her bowl of vegetable soup. Biosensors bridge the macroscopic world where our senses function with the molecular realm of the tiny disease indicators we want to detect. Spooning through the molecular soup, biosensors see and feel on a scale where we can’t.
Biosensors form this bridge by combining a molecular recognition element that specifically binds the target and a transducer that converts the recognition event into something that we can see in the macroscopic world. Today, the most frequently used biosensors for the detection of protein biomarkers are antibodies that are attached to a fluorescent tag. When added to the molecular soup, the antibody binds the target protein, if it is present, and the fluorescent tag glows to show the binding event.
This sounds simple enough, but biosensors that can be used as diagnostic tools have additional requirements. These biosensors must be inexpensive, portable, and require minimal data analysis. It must be possible to take them to the patient—in doctors’ offices or more remote medical settings—and rapidly screen for specific types of disease and cancer. In short, the biosensor must be accessible. Accessibility allows for earlier disease and cancer detection in a larger, more diverse population of patients. And early detection reduces treatment costs and gives patients a better chance at survival.
Fluorescence, the most common transducer used in biosensors today, impedes biosensor accessibility. Fluorescence-based techniques are expensive and require bulky equipment and complex, time-consuming data analysis. This is not to say that fluorescence-based biosensors are useless. In this post-genomic era, fluorescence-based biosensors have proven absolutely essential for identifying biomarkers—that is, deciding just what we find yucky in the first place. Tools such as Genomic PCR and RT-PCR utilize fluorescence to identify gene abnormalities and generate expansive maps of gene expression, respectively. From these data we can determine what proteins are indicators of the disease process in cancer patients. However, once we know our specific target, fluorescence becomes less practical for actually screening individuals. Fluorescence data are essential for informing biosensor design, but we need a new detection tool when it comes to fishing around in the soup.
Electrochemistry-based biosensors represent a rapidly growing area of research as this platform promises portability, low cost, and simple data analysis.1,2 A particularly interesting example of how a biological recognition event can be converted into an electrical signal is being developed by Professor Jackie Barton’s research group at Caltech. The Barton group’s DNA-based electrochemical biosensor is designed for the detection of transcription factors—DNA-binding proteins that regulate gene expression—as many have been identified as biomarkers for different types of cancer. Unlike fluorescence-based strategies, and most unique to this biosensor, the molecular recognition element, DNA, is also the transducer.
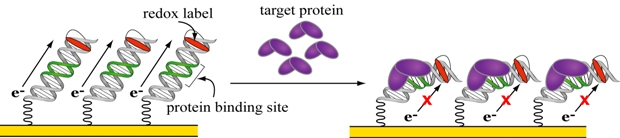
Figure 1. A DNA-based electrochemical biosensor for transcription factors.
Figure 1 illustrates the simple scheme of this DNA-based biosensor. A gold electrode is modified with a forest of DNA strands that are attached by one end to the electrode through a gold-sulfur bond. The free end of the DNA is modified with a redox label that accepts charge and thus produces an electrical current that can be measured. When the target protein binds the DNA and distorts its structure, charge cannot flow to the label and the electrical current is attenuated, indicating the binding event. The simplicity of this design arises from the role of DNA as both the recognition element and the transducer. This unique dual function is made possible by two inherent properties of DNA—a specificity for protein binding and a structurally sensitive capacity to conduct charge.
Before we can understand these two properties, it is necessary to remove the standard metaphors for DNA which frame it in a static, non-physical context. DNA is not just a sequence of letters, an instruction manual, a book with pages and pages of text in neat little rows dictating the color of our eyes and whether or not we like peas. It is a physical molecule that must function in dynamic, living organisms. We need to move and grow, fight disease, reproduce, and respond to a changing environment. The instructions that DNA holds must be accessed and controlled in real time. Tight regulation of gene expression is achieved through a complex dance of binding interactions between proteins, including transcription factors, and DNA. Participation in this dance requires DNA to have an inherently sensitive structure. This fundamental, nature-driven requirement is the foundation for the protein binding and charge conductance properties that make DNA key to this biosensor.
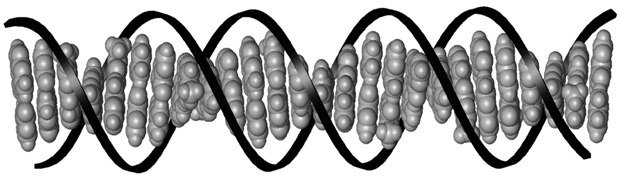
Figure 2. The stacked base pairs of DNA.3 The base pairs are shown in grey and the sugar-phosphate backbone is represented by the black ribbon.
With its letters strung together by a sugar-phosphate backbone, and curled into its characteristic double helical shape, DNA forms a landscape for transcription factor proteins to bind (figure 2). All those letters, the DNA base pairs, are individual molecules that lend subtly different physical properties to the sequences that they form. Transcription factors recognize these differences and bind at very specific DNA sequences, called binding sites. Because of this, the DNA-based biosensor can be easily modified to detect different proteins by changing the sequence of the protein binding site in the DNA strands attached to the electrode surface.
Individually, the DNA bases are flat molecules that stack on top of each other like a pile of coins (shown in grey in figure 2). Multiple studies have demonstrated the unique ability of this stack to conduct electrical charge in a process called DNA-mediated charge transport.4 Importantly to the biosensor, this capacity to conduct charge is incredibly sensitive to the proper alignment of the DNA base pair stack.5 Like putting a kink in a garden hose to stop the flow of water, when the structure of DNA is bent or distorted, charge cannot flow. Upon DNA binding, many transcription factors cause a distortion to this stack. On the DNA-based biosensor, the sequence-specific protein binding event is translated electrically as an interruption of the flow of charge to the label.
The Barton group has used this DNA-based biosensor to detect multiple proteins including the TATA Binding Protein (TBP).6,7 This transcription factor, which binds the specific site 5'-TATAAAAG-3', bends the DNA 90° upon binding, severely distorting the stack of the DNA base pairs. When TBP binds the DNA on the biosensor, the distortion cuts off the path for charge to flow from the electrode to the DNA-bound label. An attenuation of the electrical signal from the label indicates this binding event. Current work with this biosensor includes the detection of transcription factors in more complex, biological samples as well as the fabrication of multiplexed DNA electrodes for detection of an array of proteins on one device.
The development of diagnostic biosensor tools that are highly accessible will lead to earlier disease and cancer detection for more people, reducing medical costs and improving the prospects of patient recovery. As we strive toward this goal, there is still a great need for creative solutions. DNA as a combined recognition element and transducer is one such solution to this challenge in which our fingers and eyes alone can’t determine if the soup contains peas. In this case DNA acts as an exquisitely sensitive extension of our fingers into the molecular realm. Poking into the soup, the DNA can feel the proteins for us.
References:
1. Drummond, T. G., Hill, M. G., Barton, J. K. Electrochemical DNA Sensors. Nat. Biotech. 10, 1192-1199 (2003).
2. Sadik, O. A., Aluoch, A., Zhou, A. Status of biomolecular recognition using electrochemical techniques. Biosensors and Bioelectronics 24, 2749-2765 (2009).
3. Figure adapted from: Delaney, S., Barton, J. K. Long range DNA charge transport. J. Org. Chem. 68, 6475-6483 (2003).
4. Wagenknecht, H., ed. Charge Transfer in DNA: From Mechanism to Application. Weinheim: Wiley-VCH, (2005).
5. Kelley, S. O., Holmlin, R. E., Stemp, E. D. A., Barton, J. K. Photoinduced electron transfer in ethidium-modified DNA duplexes: dependence on distance and stacking. J. Am. Chem. Soc. 119, 9861-9870 (1997).
6. Boon, E. M., Salas, J. E., Barton, J. K. An electrical probe of protein-DNA interactions on DNA-modified surfaces. Nat. Biotech 20, 282-286 (2002).
7. Gorodetsky, A. A., Ebrahim, A., Barton, J. K. Electrical detection of TATA binding protein at DNA-modified microelectrodes. J. Am. Chem. Soc. 130, 2924-2925 (2008).






