By understanding how proteins fold, and what structures they are likely to assume in final form, researchers are then able to move closer to predicting their function.
Modeling protein folding on computers has existed for decades but McGill researcher Jérôme Waldispühl of the McGill Centre for Bioinformatics and his team has developed algorithms that can examine a protein's fundamental chemical properties and then scan a number of possible protein shapes before predicting the final form that the protein is likely to take. They call it tFolder.
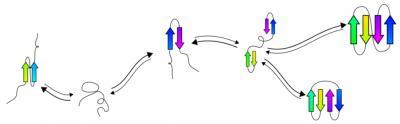
The process of predicting protein folding. Credit: tFolder
tFolder results have been impressive, shortening classical techniques for predicting protein folding pathways from hundreds of thousands of CPU hours to compute the folding dynamics of 40 amino acids proteins, down to 10 minutes on a single laptop, a coarse-grained representation of the folding pathways of a protein with 60 amino acids.
Waldispühl and his students continue to work on the tFolder algorithm to improve its success rate at predicting protein folding with broader categories of proteins including some that are important in DNA-binding. The research was recently presented at the 15th Annual International Conference in Research in Computational Molecular Biology (RECOMB 2011).





Comments