Aging is a complex phenomenon and the mechanisms underlying aging are yet to be explained. What researchers do know is that there is a clear relationship between aging and calorie intake. For example, mice fed with half the calories they usually eat can live 40 percent longer. How does this work?
As part of the study, Université de Montréal Biochemistry Professor Luis Rokeach and his student Antoine Roux discovered to their surprise that if they removed the gene for a glucose sensor from yeast cells, they lived just as long as those living on a glucose-restricted diet. In short, the fate of these cells doesn't depend on what they eat but what they think they're eating.
There are two obvious aspects of calorie intake: tasting and digestion. By the time nutrients get to our cells there is an analogous process: sensors on the surface of the cell detect the presence of, for example, the sugar glucose and molecules inside the cell break down the glucose, converting it to energy. Of these processes, it is widely thought that the by-products of broken down sugars are the culprits in aging. The study by Rokeach and Roux suggests otherwise.
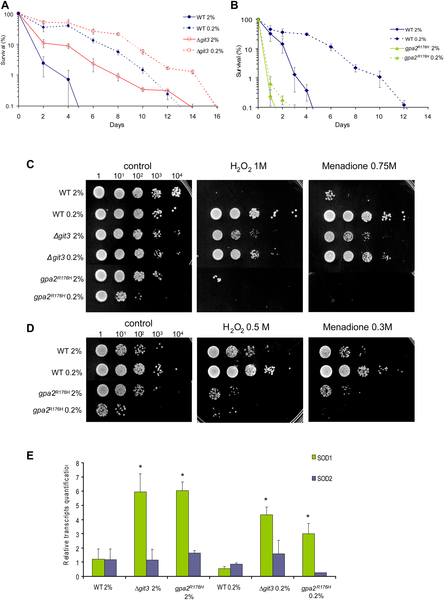
Longer chronological life span correlates with higher mitochondrial respiration and increased oxygen stress resistance, except with GpaR176H strain where loss of survival correlates only with weaker oxidative stress resistance. (A) CLS of <Δgit3 grown in 2% and 0.2% glucose as compared to wild type. (B) Chronological life span of gpa2R176H grown in 2% and 0.2% glucose. (C) Oxidative stress resistance measured in WT, Δgit3 and gpa2R176H after short treatment of H2O2 and menadione. Cells were collected at day 1 of stationary phase and submitted to oxidative stressors before plating. (D) Oxidative stress resistance measured in WT and gpa2R176H at weak dose of H2O2 and menadione. (E) Transcript levels of cytosolic and mitochondrial superoxide dismutase (SOD1 and 2) normalized to wild type cells as measured by quantitative RT-PCR. Total RNAs were isolated in cells collected at day one of stationary phase. The transcript levels were normalized on two constitutively expressed genes. Standard deviations were calculated on three independent experiments; student test was done comparing with wild type at glucose 2%. *, p<0.01. Source: Roux AE, Leroux A, Alaamery MA, Hoffman CS, Chartrand P, et al. (2009) Pro-Aging Effects of Glucose Signaling through a G Protein-Coupled Glucose Receptor in Fission Yeast. PLoS Genet 5(3): e1000408. doi:10.1371/journal.pgen.1000408
To understand aging, Rokeach and Roux in collaboration with Université de Montréal Biochemistry Professors Pascal Chartrand and Gerardo Ferbeyre used yeast as a model organism. At a basic level, yeast cells are surprisingly similar and age much like human cells, as well as being easy to study.
The research team found that the lifespan of yeast cells increased when glucose was decreased from their diet. They then asked whether the increase in lifespan was due to cells decreasing their ability to produce energy or to the decrease in signal to the cells by the glucose sensor.
The scientists found that cells unable to consume glucose as energy source are still sensitive to the pro-aging effects of glucose. Conversely, obliterating the sensor that measures the levels of glucose significantly increased lifespan.
"Thanks to this study, the link between the rise in age-related diseases and the over-consumption of sugar in today's diet is clearer. Our research opens a door to new therapeutic strategies for fighting age-related diseases," says Rokeach.
Citation: Roux AE, Leroux A, Alaamery MA, Hoffman CS, Chartrand P, et al. (2009) Pro-Aging Effects of Glucose Signaling through a G Protein-Coupled Glucose Receptor in Fission Yeast. PLoS Genet 5(3): e1000408. doi:10.1371/journal.pgen.1000408




Comments