In a protein coding sequence of DNA, three subsequent nucleotides form codons (see figure 1), which, in turn, form amino acids that will eventually give rise to a protein. Some of these codons, however, are synonymous, meaning that one amino acid can be encoded by several different codons. When an amino acid is preferentially encoded by one of several potential synonymous codons, it is called codon bias.
Figure 1: Example of a codon. (GCA codes for alanine, which is also encoded by the synonymous codons GCT, GCT and GCG.)
(Source: Georgia Southern University, Department of Biology.)
A study in PLoS ONE has taken a look at potential macro-evolutionary effects of such codon bias. To investigate this, the researchers compared genome-wide codon bias in six species that occupy important positions in the Eukaryotic Tree of Life, namely Saccharomyces cerevisiae (yeast), Arabidopsis thaliana (thale cress), Caenorhabditis elegans (roundworm), Gallus gallus (chicken), Pan troglodytes (chimpanzee) and Homo sapiens.
For each protein coding sequence in these six genomes, 20 random sequences that encode the same amino acid sequence were generated. Based on this, the codon bias could be characterized. Most of the sequences in these six model organism displayed some extent of codon bias (see figure 2).
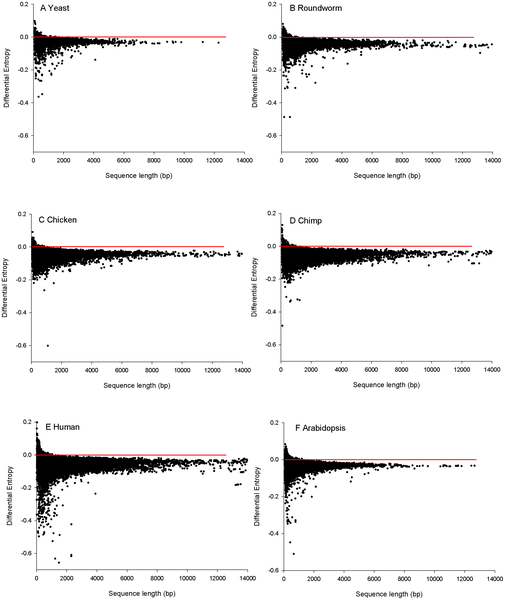
Figure 2: Protein coding sequences for each species, with their Differential Entropy. Negative values mean that the sequence is more regular than expected. So, everything beneath the red line exhibits some extent of codon bias.
(Source: Hudson et al.; 2011)
Through examining the relationship between codon bias and protein function, the research team noted that:
… we detected very strong signals suggesting that entire batteries of functionally-related genes have been subject to selection for extreme differential codon bias in a lineage-specific manner.
This is illustrated in table 1.
Table 1: The 5 most extreme functional enrichments for each species.
(Source: Hudson et al.; 2011)

The authors, however, are careful enough to mention a caveat:
… we have interpreted extreme differential codon bias in energetic terms, codon bias can also arise for other reasons – some of which are adaptive and some of which are neutral. For example, codon bias may ariseas a simple artifact of history, following duplication or some other expansion events from a smaller piece of ancestral sequence.
However, they stand firmly behind their approach:
Fundamentally, however, it seems unlikely that these alternative explanations can adequately account for the consistent functional enrichment scores detected. Our explanation rests on the observed enrichment for whole pathways or processes, not individual molecules. For example, non-energetic hypotheses could potentially explain why the LCE proteins possess extreme codon bias in mammals. But it stretches credibility that these same non-energetic hypotheses also explain the KRTAP gene sequences, given the two groups of sequences have an independent evolutionary trajectory.
Overall, they conclude:
We have systematically quantified codon bias in several key eukaryotes. In doing so, we have identified lineage-specific patterns of codon bias that have not previously been reported. Some of these only became apparent through comparisons between species. Our working hypothesis is that patterns of extreme codon bias highlight molecules and pathways from a particular lineage that have been given energetic priority (assuming bias towards preferred codons) through natural selection. These patterns identify genes and gene families unique to, or having particular relevance in, a given lineage (such as hair in mammals, and cell walls in plants). Our hypothesis is supported by functional enrichments for entire pathways or processes, not merely individual molecules. These enrichments are built on observations of cohorts of DNA sequences that possess independent evolutionary histories and quite different messenger RNA characteristics.
Reference
Hudson, N.J.; Gu, Q.; Nagaraj, S.H.; Ding, Y.-S.; Dalrymple, D.P. and Reverter, A. (2011). Eukaryotic Evolutionary Transitions Are Associated with Extreme Codon Bias in Functionally-Related Proteins. PLoS ONE 6(9):e25457. doi:10.1371/journal.pone.0025457. (Click here for the article.)




Comments