We have been involved with docking studies before. Of course, Tack Kuntz at UCSF is a Dock pioneer. In general, Dock studies have identified some low uM inhibitors of falcipains, but it has been hard to move beyond this. However, these studies were based on models of FP structures, based on other papain-family enzyme structures. Now we have structures determined for the two key targets, falcipain-2 (the link that you sent) and falcipain-3 (not yet published, I am not sure if coordinates are yet available). These structures are from a structure group at UCSF. The key question is whether you will be offering a new approach compared to that used previously. It appears that the answer is yes, as you will have access to solved structures, and also to new chemistry methods. From our part, we’ll be happy to do the small number of screens that you envision. 5-10 compounds is quite trivial. I mention to all collaborators that we have many collaborations with chemists interested in protease inhibitors as antimalarial drugs. Our major collaboration is with a group at GSK, and this project is well advanced using a traditional big pharma approach. We have academic collaborations with chemists in the US, Germany, Portugal, Venezuela, Cameroon, India, and South Africa (not only regarding protease inhibitors). I make it a point to be very open regarding the different collaborations but, of course, to respect each group’s confidentiality concerning results. For a small set of compounds, we can do screening at any time. For larger projects, we can talk about the path forward. I’ll be in touch, and please let me know any time when you have compounds that you would like to screen or if you have any questions
Falcipain Collaboration To Fight Malaria
Thanks to Barry Bunin of Collaborative Drug Discovery, we now have a collaborator who will run assays on the compounds from our CombiUgi project. We'll be using our account on CDD to manage the activity results.
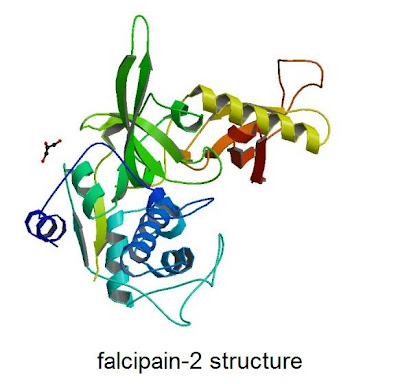
Phil Rosenthal from UCSF has agreed to run assays on the inhibition of falcipain-2, an enzyme used by the malaria parasite to digest hemoglobin. As described in UCSF magazine, the Rosenthal group discovered the enzyme and have developed an assay.
That means we'll have to do docking of our 500K library on falcipain-2. The PDB file is available here.
This is very good news for demonstrating how real-time Open Science can be used from drug design to synthesis to testing.
Here is an email from Phil to clarify some more details:






Comments