A new genomic characterization of E. coli strains isolated directly from diabetic foot ulcers across multiple continents may help explain why some infections become difficult to treat and lead to severe, even life-threatening, outcomes. The team analyzed whole-genome sequences from 42 E. coli strains isolated from infected diabetic foot ulcers in patients from Europe, Africa, Asia, and the Americas and sequenced the complete DNA of each bacterial strain.
They were then able to to compare genetic differences between strains, identify genes linked to antibiotic resistance, and pinpoint factors that contribute to disease severity. Around eight percent of the strains were multidrug-resistant or extensively drug-resistant, meaning they are resistant to multiple or nearly all available antibiotics and the E. coli strains were highly diverse. The bacteria belonged to many different genetic groups and carried a wide range of genes linked to antibiotic resistance and disease.
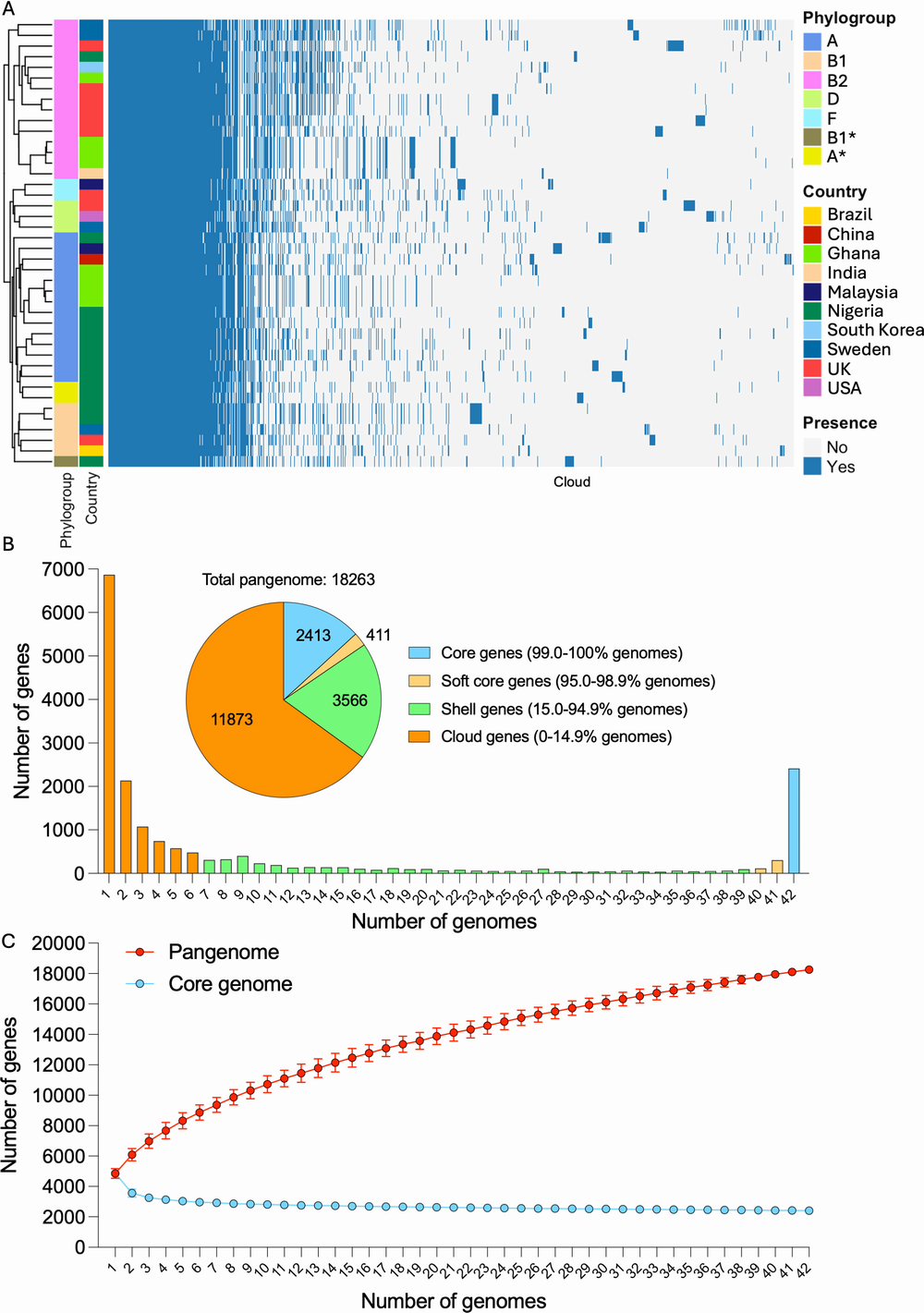
(A) Gene presence/absence heatmap. Genomes were annotated using Prokka, and the pangenome was reconstructed using Roary. Strain clustering is based on patterns of presence/absence. Phylogroups and countries of origin are labeled in different colors. (B) Gene frequency histogram plot, representing the number of genomes sharing different numbers of genes. Singletons are displayed at the far left, while core genome genes are represented at the far right. The inset pie chart represents the overall number of genes within core, soft core, shell, and cloud genomes. (C) Pangenome and core genome accumulation curves. Data are represented as average ± SD of 100 reinteractions performed with different randomized orders of genomes.
The finding affirms that that there is no single type of E. coli responsible for diabetic foot infections, and distinct lineages were independently capable of adapting to the diabetic foot environment. Next, they identified the resistance mechanisms and virulence traits they carry, which helps explain why some diabetic foot infections are particularly difficult to treat or can progress rapidly to severe illness.
Citation: Ajumobi V, Tahir Z, Hayes P, McCormick A, Torraca V. 0. Population structure, antimicrobial resistance, and virulence factors of diabetic foot-associated Escherichia coli. Microbiol Spectr 0:e02837-25. https://journals.asm.org/doi/10.1128/spectrum.02837-25






Comments