Many computational biologists are interested in taking gene expression data, and using that data to computationally infer the underlying regulatory network that controls the observed pattern of gene expression.
Why? Because doing the experiments to determine the structure of these regulatory networks is hard; if we could use more easily obtained data to reliably tease out the network structure, we'd be able to quickly characterize networks in unexplored cell types or in poorly studied microbes.
The problem when making these network-inferring algorithms is knowing whether you're getting the right answer - the main justification for these algorithms is that determining the right answer is hard to do experimentally, so what researchers need is a set of known regulatory networks on which to test their computational methods.
A group from the University of Naples, Italy, built a synthetic, 'gold standard' regulatory network on which computational biologists can test their network-inference programs. This toy network, consisting of five transcription factor genes wired up into a set of feedback loops, is an interesing idea in principle. The piece was accompanied by a commentary titled "Systems Biology Strikes Gold."
In practice, things aren't so glittery: this toy network basically does nothing interesting. As far as I can tell, the researchers arbitrarily wired these genes together without giving much thought to designing something with non-trivial dynamics. As a result, when they switch the network on (by adding galactose to cultured yeast - the network was made inside of a yeast strain), not much happens. They built a mathematical model of this network, and they sort-of matched some bumpy, largely flat lines.
You can see this in the figure below. The x-axis is time, the y-axis is gene expression, the dots are the experimental data (with the standard error shown by the dashed lines), and the model prediction is the solid colored line.
The big bump in the GAL4 panel below, the most dramatic thing that happens, was actually an experimental artifact caused during the transfer of yeast from glucose to galactose, and had to be added to the model with a fudge factor.
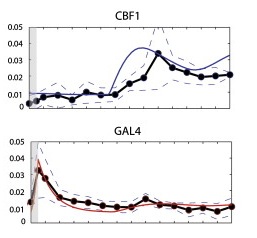
When the researchers tested various network reconstruction algorithms on this very sedate synthetic network, none of the programs got it right. You have a simple network with five nodes and eight edges, and no algorithm got it completely right. Some didn't even do any better than random chance.
To be fair, this network didn't really do anything, so these algorithms were trying to say something about almost nothing. It's possible that the programs would do better on a more dynamic system.
This is also a sign that network inference is a hard problem. The best results of the state-of-the-art programs aren't yet good enough.
In other words, experiments aren't obsolete yet.





Comments