Increasing resistance of pathogens to antibiotics is an important issue. The hunt for new antibiotics/antimicrobials is on. For example, the 10X’20 Initiative aims to develop 10 new antimicrobial drugs by 2020. But where will these new drugs come from?
The recent efforts in sequencing marsupial genomes might prove helpful. Since the young of marsupials and monotremes are relatively little developed at birth, and have to survive outside of the sterile environment of the uterus, their immune system has to be capable of dealing with potential pathogens in their environment.
A new study, published in PLoS ONE, went looking for antimicrobial peptides in the genomes of several non-placental mammals. Specifically, they searched for cathelicidin genes, one of the most well-known antimicrobial gene families. The hunt for genes resulted in a great diversity of cathelicidin genes in the genomes of the tammar, the opossum and the platypus (see figure 1). These genes are quite old, diverging from each other about 211 million years ago. In the eutherian lineage, many of these genes were lost, likely due to the evolution of a longer gestation period and young with a more competent immune system. The recent (about 53 mya) gene duplications in sheep, cows and pigs (see figure 1) were likely driven by an increased pathogen pressure in herd animals.
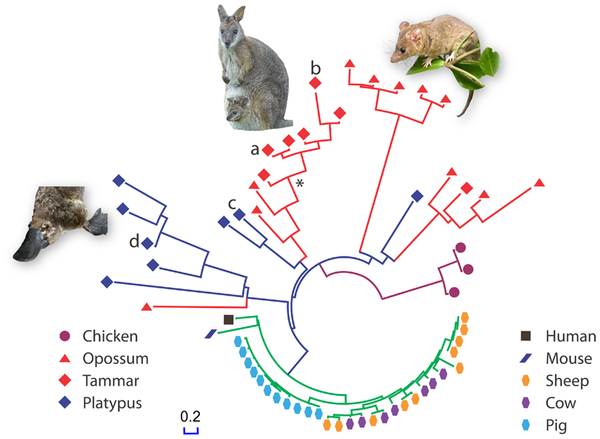
Figure 1: Phylogenetic tree showing the diversity of antimicrobial peptides in non-eutherian mammals.
(Source: Wang et al., 2011)
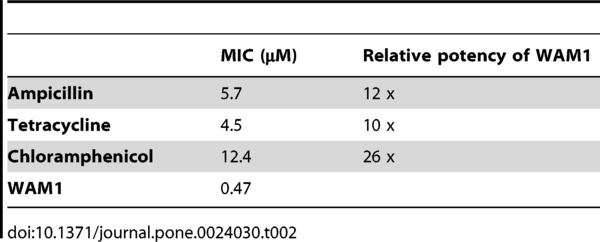
Four of the cathelicidin peptides (two of the tammar: WAM1 and WAM2, and two from the platypus: PAM1 and PAM2) were synthesized and tested. All four were found to be very effective against several pathogens when compared to the human cathelicidin (see table 1). WAM 1 seems to be “remarkably potent”, being 3 – 80 times more successful than the human peptide, and 10 to 26 times more effective than three antibiotics (see table 2).
Table 1: Antimicrobial activity of four peptides compared to a hypothetical ancestral WAM and LL37, the human peptide. (MIC = minimum inhibitory concentration)
(Source: Wang et al., 2011)
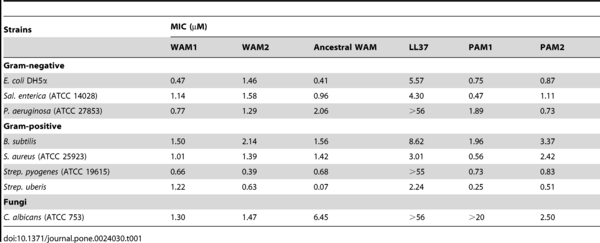
Table 2: Potency of WAM1 compared to three antibiotic drugs.
(Source: Wang et al., 2011)
The authors conclude:
The dwindling numbers of therapeutic options for bacterial ‘superbugs’ necessitates the development of novel antimicrobials. The development of microbial resistance against naturally occurring antimicrobials peptides is rare, and our data suggest that the potent antimicrobial peptide gene expansions in the platypus and tammar genomes are good candidates for discovering natural peptides to fight multidrug resistant microbes. The strong antimicrobial activity, salt insensitivity and low haemolytic activity further suggest that marsupial and monotreme cathelicidin peptides have the potential to provide novel therapeutic antibiotics.
Reference
Wang, J.; Wong, E.S.W.; Whitley, J.C.; Li, J.; Stringer, J.M.; Short, K.R.; Renfree, M.B.; Belov, K. and Cocks, B.G. (2011). Ancient Antimicrobial Peptides Kill Antibiotic-Resistant Pathogens: Australian Mammals Provide New Options. PLoS ONE. 6(8): e24030. doi:10.1371/journal.pone.0024030. (Click here for the article)




Comments