Earlier this year, a DNA computer was reported that could calculate square roots. A little later, a neural network was constructed out of ‘the stuff of life’. These advances strongly hinted at the possibility of using biochemicals in order to do computational operations. Now, a new study, published in Nature Communications, presents another step towards this goal.
A research team from Imperial College in London has created biological logic gates in Escherichia coli. To achieve this, the team used genetics to devise a logical AND gate (see figure 1). In short, environmental signals can bind to promoters and result in the activation of two genes, hrpR and hrpS. When the products of both genes are present, they bind to the activator sequence of another gene. The result of this is that a closed transcription complex is remodeled into an open one, leading to the transcription of the third involved gene, which is a reporter gene (gfp) that enables the researchers to see that their inputs result in the desired output.

Figure 1: A biological AND gate. Environmental signals (I 1 and 2) bind to a promoter (P 1 and 2), activating genes (hrpR and hrpS respectively). The products of these genes (R and S) bind to an activator sequencr, thereby changing the form of the sigma54-RNAP transcritption complex, which allows the transcription of the gfp gene.
(Source: Wang et al., 2011)
Furthermore, the authors show that their biological logic gates can be combined to form more complex components. They combined a NOT gate with an AND one to get a NAND gate (see figure 2). Here, the output of the AND gate is used as input for the NOT one, resulting in a NAND logical gate.
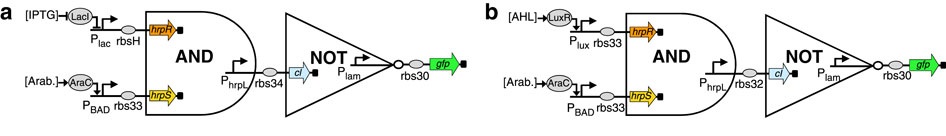
Figure 2: Two potential NAND gates.
(Source: Wang et al., 2011)
The authors mention a few features of their gates that make them potentially suitable to operate in biological systems, perhaps resulting in microscopic biological sensors that can detect and fight off certain pathogens, or combat cancer or toxins:
Because of modularity, the AND and NAND gates can be reconnected to different sensor inputs to integrate multiple environmental and cellular signals and thus be applied to identify a specific combinatorial condition, like the cancer microenvironment. The transcription-based AND gate was shown to be robust with functionality in a range of different tested contexts. Moreover, the signal response surfaces of the AND and NAND gates show the sigmoid response property for both inputs, largely due to the high cooperativity of the two transcriptional activator proteins. The sigmoidal response filtering property can suppress the analogue noise in biomolecular logic systems and increase signal robustness, enhancing the error tolerance capability and the scalability of the logic gates. Having a similar role as their electronic counterparts, the engineered AND and NAND gates are the fundamental components in designing biologically based digital systems to control cell signalling and can be easily applied in different contexts, owing to their modularity and orthogonality, to implement human-designed intra- and extra-cellular control functions.
Before this happens, further research is, of course, required. Nevertheless, this represents another step forwards towards such goals. The authors conclude:
As demonstrated in the construction of the genetic logic gates, the diverse natural building blocks in the myriad of prokaryotic and eukaryotic species provides a rich source from which to engineer orthogonal devices with a variety functions.
Will be continued…
Reference
Wang, B.; Kitney, R.I.; Joly, N. and Buck, M. (2011). Engineering modular and orthogonal genetic logic gates for robust digital-like synthetic biology. Nature Communications 2 article 508. Published online 18 October. Doi:10.1038/ncomms1516. (Click here. Open access. Kudos for that, Nature Communnications)





Comments