Some genes appear to have an effect on lifespan. This shouldn’t be too surprising news. But now, a research team from Stanford has shown that there are epigenetic effects on longevity as well. Using the nematode Caenorhabditis elegans, a beloved model organism in aging research, they have shown that some changes in chromatin states in a parental generation can affect the lifespan of their descendants.
More specifically, they looked at the regulatory complex H3K4me3, which is composed out of ASH-2, WDR-5 and SET-2, and responsible for adding methyl-groups to the H3 histone. It has been shown that deficiencies in this complex extend lifespan. So, the researchers proceeded by ‘knocking out’ each of the components of this complex separately and determine whether or not this had an effect on the lifespan of the little worms.
The disturbance of each of the components significantly increased lifespan. Not only of the worms in which the components were disabled, but, and this is the important part, also in their descendants up to three generations later (the increase ranged from 20 to 30%) (see figure 1). So, even without changes in the DNA, the descendants somehow inherited the epigenetic markers of the parental generation. These markers are thought to be ‘reset’ each generation, and yet some epigenetic markers are transferred to the next generation. How exactly this happens is one of the big questions in epigenetics.
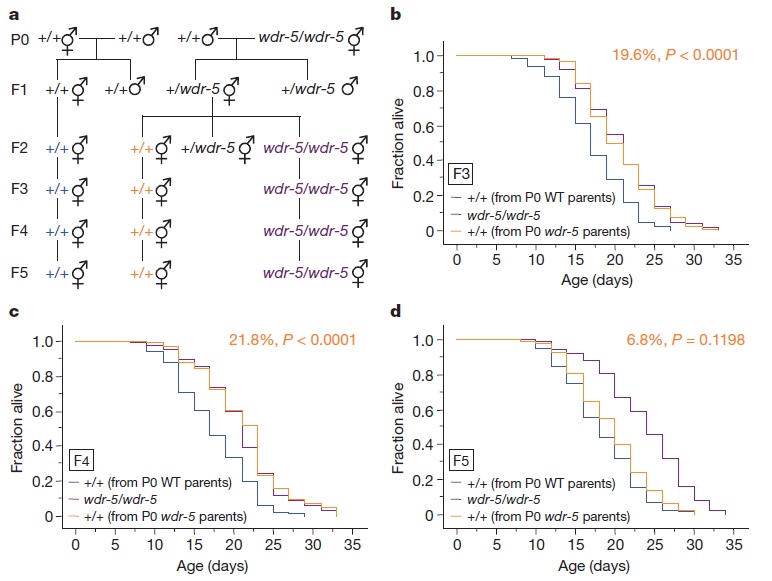
Figure 1: (a) Breeding schedule of the worms, where +/+ is the wild-type and wdr-5/wdr-5 the worms where WDR-5 is 'knocked out', (b), (c) and (d) represent the lifespan of the descendants after 3, 4 and generations respectivley. (Similar results were obtained for ASH-2 and SET-2.)
(Source: Greer et al., 2011)
This study shows that even traits as complex as lifespan can be significantly influenced by epigenetic markers.
The authors conclude:
Our study provides the first example of epigenetic inheritance of longevity. Histone methylation marks and DNA methylation are generally, but not always, erased between generations with epigenetic reprogramming. Our observations are consistent with the notion that H3K4me3 at specific loci may not be completely erased and replenished. … As the ASH-2 H3K4me3 regulatory complex is conserved from yeast to humans, manipulations of this complex in parents might have a heritable effect on longevity in mammals.
To test this, the lab is currently testing this in African killfish and mice. While this might not be as relevant in vertebrates as in nematodes, it’s certainly worth taking a look, isn’t it?
Reference
Greer, E.L.; Maures, T.J.; Ucar, D.; Hauswirth, A.G.; Mancini, E.; Lim, J.P.; Benayoun, B.A.; Shi, Y. and Brunet, A. (2011). Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature. Published online 19 October. doi:10.1038/nature10572.





Comments