Jumping genes, or more technically, transposons (see figure 1), are sequences of DNA that can move around the genome and find themselves a new place. In eukaryotic DNA, these jumping genes can constitute a sizeable portion of the genome (up to 50% of the human genome is made up out of active transposons and the remains of former ones that became inactive).
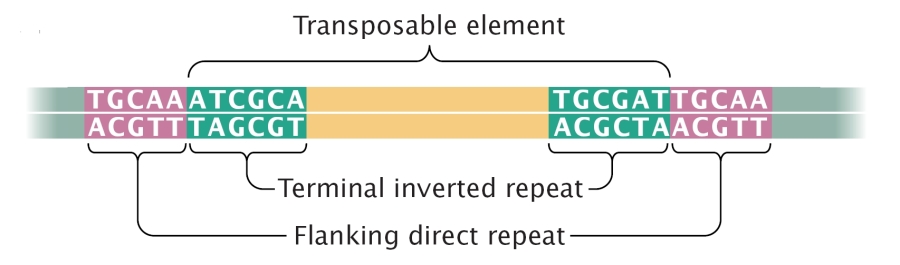
Figure 1: General structure of a transposon.
(Source: Scitable, by Nature Education)
Beyond being a mystery in themselves, these bits of moving DNA (and their remnants) are remarkably prevalent in chromosome regions that are the last ones to replicate.
Now, a new study might have found the reason why. The study focused on one particular transposon, known as the P element, in the genome of Drosophila melanogaster. This P element has only been around for about 80 years, but has quickly spread. The research team looked at a database containing more than 50,000 genomic sites where the P element has inserted.
They found that the P element was quite selective in finding its insertion place. What many of these insertion places have in common is that these sites have the ability to function as starting sites of DNA duplication. This association between P element insertion and genome duplication suggests that this transposon can coordinate its movement with DNA duplication.
The research team proposes that the P element (and maybe other transposons as well) use this to spread more rapidly through the genome. The elements would transpose after being replicated, and insert themselves into a site that hasn’t replicated yet, allowing them to replicate twice instead of just once in a single genome duplication cycle.
This also means that, if these elements get a late start, only the last still-to-be-duplicated chromosome segments will provide them a chance for a double duplication. As a consequence, these regions should be rich in transposon elements. An important note here is that other transposons, such as piggyBac and Minos, do not insert themselves near replication origins. The mechanism is thus far from universal.
Nevertheless, this is an intriguing finding. In the words of one of the authors:
By gaining insight into one specific transposon’s movements, we may have begun to glimpse mechanisms that have profoundly influenced genome evolution for nearly all animals.
References
Spradling, A.C.; Bellen, H.J. and Hoskins, R.A. (2011). Drosophila P elements preferentially transpose to replication origins. Proceedings of the National Academy of Sciences. Published online before print, 6 September 2011. doi:10.1073/pnas.1112960108.
Carnegie Institution for Science, News.





Comments