Researchers at the European Molecular Biology Laboratory's European Bioinformatics Institute (EMBL-EBI) and their colleagues say this visual language should make it easier to exchange complex information, so that models are accurate, efficient and readily understandable. The new standard, called the Systems Biology Graphical Notation (SBGN), is published today in Nature Biotechnology.
Researchers use standardized visual languages to communicate complex information in a way that it is unambiguous and easy to understand. Such standard graphical representations are common to many scientific fields.. But biology still lacks a standardised notation that describes all biological interactions, pathways and networks, even though the discipline is dominated by graphical information.
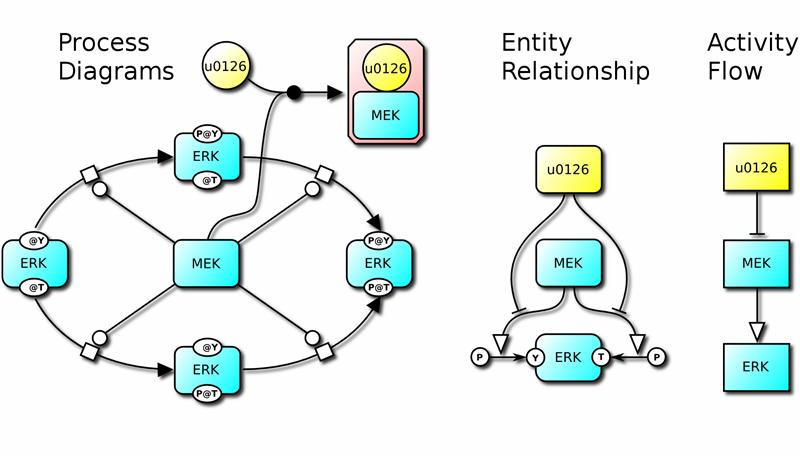
Graphic showing the catalysis of ERK (extracellular signal-regulated kinase) phosphorylation by the kinase MEK in the growth factors signalling pathway. This is illustrated by the three SBGN diagrams: process, entity relationship and activity flow. Copyright EMBL.
The SBGN project was launched in 2005 as a united effort to specifically develop a new graphical standard for molecular and systems biology applications. The project was initiated by Hiroaki Kitano (Systems Biology Institute, Tokyo, Japan) and coordinated by Nicolas Le Novère (EMBL-EBI, Hinxton, UK) and Michael Hucka (California Institute of Technology, Pasadena, USA). The team comprises biochemists, modellers and computer scientists who have developed the SBGN in collaboration with the systems biology research community.
Le Novère said: "In the genomics era, especially since the emergence of high-throughput technologies, there have been massive increases in the amount of biological data. We believe that the SBGN will make it easier for researchers to understand each other's models and to share this data more effectively. This will benefit systems biologists working on a variety of biochemical processes, including gene regulation, metabolism and cellular signalling".
To ensure that this new visual language does not become too vast and complicated, the researchers decided to define three separate types of diagram that complement each other, describing molecular processes, relationships between entities and links among biochemical activities. This simplicity, combined with the extensive involvement of the community of researchers that will use SBGN, should ensure that the notation is rapidly adopted and widely used.
Article: Le Novère N, Hucka M, Mi H, Moodie S, Shreiber F, Sorokin A, Demir E, Wegner K, Aladjem M, Wimalaratne S, Bergman FT, Gauges R, Ghazal P, H Kawaji, Li L, Matsuoka Y, Villéger A, Boyd SE, Calzone L, Courtot M, Dogrusoz U, Freeman T, Funahashi A, Ghosh S, Jouraku A, Kim S, Kolpakov F, Luna A, Sahle S, Schmidt E, Watterson S, Goryanin I, Kell DB, Sander C, Sauro H, Snoep JL, Kohn K, Kitano H, 'The Systems Biology Graphical Notation', Nature Biotechnology (2009), 27






Comments