If you had one hundred unlabeled DNA samples, taken from people all around the world, could you use that DNA to determine where the original donors came from?
With major improvements in genotyping technology, geneticists are now getting better and better at this game, and a recent paper in Science reports the largest study to date of human genetic diversity: 650,000 genetic differences scrutinized in nearly 1000 different individuals from 51 different populations.
Studies like this one lay important groundwork to help us understand how human genomes differ around the world, how differences in our genes and environments together make us healthy or sick, and how very ancient migrations led to the structure of today's human populations around the globe.
A group at Stanford University used Illumina's BeadArray chips to determine the identity of the DNA base at 650,000 different places where humans vary genetically, known as single nucleotide polymorphisms, or SNPs - essentially where just one 'letter' of DNA differs in people. They did this for DNA samples from nearly 1000 people from around the world. Where did they get all these samples? From a remarkable DNA bank: a collection of self-perpetuating white blood cells taken from 1000 volunteers, the Human Genome Diversity Panel.
Researchers collected blood from these volunteers, isolated what are called B cells, and then immortalized these cells using the Epstein-barr virus. This created what are called cell lines, self-perpetuating cultures of cells that can then be studied by hundreds of different labs around the world. With frozen blood samples, some day you run out and you have to find new volunteers; with cell lines, you never run out of DNA.
Some labs are even studying these immortal cell lines themselves; for example, you could see how different cell lines respond to drugs, and then trace those drug-response differences to genetic differences in the cells. The people who donated these blood samples now have far more of their cells outside of their bodies than inside, spread out in research labs around the world.
So how well did the Stanford group do at reconstructing human geography from pure genetics? They took their genetic data, and without looking at where the DNA samples had come from, assumed that their samples originated from seven ancestral human populations (they chose seven to match the number of broad regions they were examining, but they also did the analysis with numbers other than seven). The problem was now to see, without peeking at the answer, which individuals came from which ancestral population.
The researchers put together a computer program which identified groups genetic differences that tend to stick together: if your genome has an 'A' here, a 'T' over there, another 'A' back there, a 'G' up here, then it looks like you came from ancestral population 1. Someone else with a another group of genetic variants possibly came from ancestral population 4. To further complicate things, some people have a little population 1, and a little population 4, and maybe even some 7.
But when you sort it all out, it looks like this:
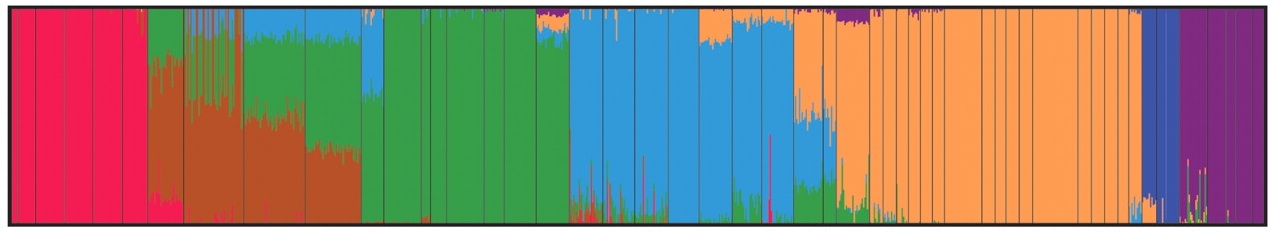
In this figure, there are 938 vertical stripes (too small to make out individually) - each stripe is an individual, and the color of the stripe represents which ancestral population that individual's genome says they came from. You can see some mixed stripes, individuals who are genetically mixed (and presumably from populations that are genetically mixed).
Here comes the impressive part: when you add geographical labels to the figure, you find that the genes match up with the geography:
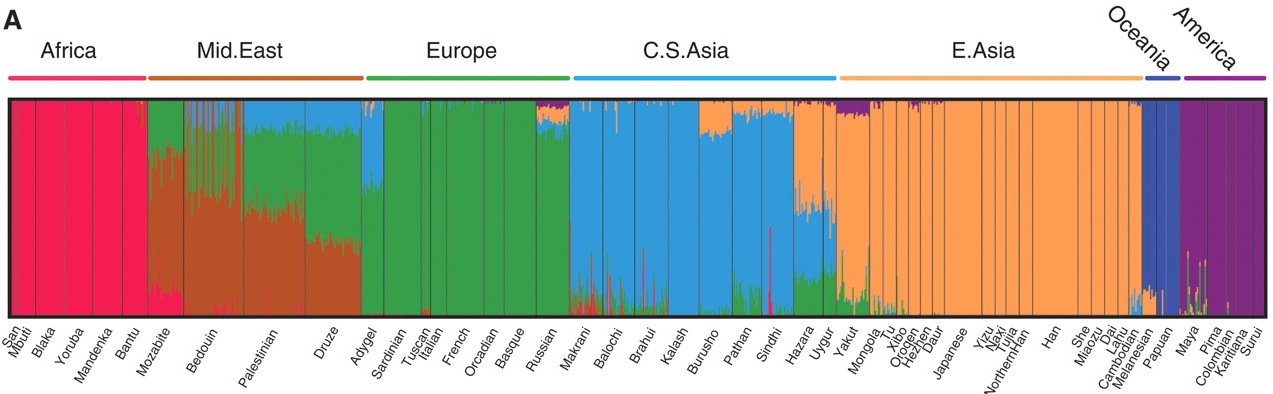
Researchers have done things like this before, but never in such detail. You can see that the Levantine Druze population (or at least the individuals included in this study) has a mixture of Middle Eastern, European, and Central Asian genetic variants, while the Hazara (think The Kite Runner) appear to have European, Central Asian, and East Asian ancestry.
Paradoxically, we can use genes to divide up humans into ancestral populations, but most of our genetic differences are within populations, not between them. That means that while you can use a small number of key differences to distinguish my genome from that of a Hazara, on the whole, I am likely to not be any more different genetically from a Hazara, in terms of sheer number of SNP differences, than I am from a random individual from Europe (with whom I share an ancestral population). Only about 10% of our genetic differences delineate human geographical populations; in the other 90% we can differ even from those people who share our geographical ancestry.
Where does this research go next? First, it goes to hundreds of different labs around the world working with the immortalized B cell lines, who can now take advantage of the thorough genetic data that the Stanford group has made available. These groups are looking for the genetic differences underlying everything from our response to drugs to the ability of B cells to react to immune system signals.
This research also paves the way for further research into deep human history. Where did the world's different populations - those that populated the Americas, Oceania, the Middle East, all ultimately traceable back to Africa - come from? How large were they, and when did they migrate? What can their genes tell us about the ancient environmental pressures they faced, or about the distinct disease patterns among ethnic groups today? With the powerful genotyping technology available today, we can now approach those questions with a confidence that would have been foolhardy just 25 years ago.





Comments