Aside from wondering if you’re going to die soon, I’m almost positive the second and third questions on your mind are; what are the molecular mechanisms that make Influenza A so nasty? And then maybe, how the heck did Wilbur the pig, a distantly related creature, manage to spread his cooties to humans?
Influenza A infects approximately 10% of the population in the US each year and causes approximately 35,000 deaths annually. That's arguably a good year considering Influenza A also makes a recurring appearance every 10 to 15 years as a highly virulent form that is responsible for epidemics.
So what is it about Influenza A that makes it a top candidate for pandemics? The primary answer lies in its genomic layout.
Influenza A is a RNA virus and in general, RNA viruses have a very high rate of mutation, a short generation time and yield a high production of virions after replication in the host’s cells. However, influenza A’s most advantageous feature is its segmented genome which consists of eight different linear RNA strands. These strands can combine with each other and create novel genotypes—an important mechanism for extremely rapid adaptation to new hosts.
The major advantage conferred by a segmented genome comes into play during coinfection which is when a cell becomes infected by two different viral strains at the same time. During coinfection, different segments of each virion can recombine, thus creating a new viral strain emergence that is sometimes even more detrimental than the original parent viral strains. Furthermore, viral strains from different species can also recombine with each other.
When two different species’ influenza strains (for example pig and human) recombine, the novel viral strain that emerges potentially has a better chance of evading the immune system and persisting in the host organism than the pre-existing viral strains. In other words, the host organism’s immune system doesn’t have antibodies for viruses indigenous to other species and thanks to the original virus that is indigenous to the host, molecular features necessary for “optimal” host infection are retained.
The emergence of a viral strain that is capable of evading the host's immune system as well as transmits and replicates effectively, is precisely the recipe for a potential pandemic.
Every 10 to 15 years, influenza undergoes a dramatic genetic change (antigenic shift) which as described, is responsible for epidemics/pandemics. In between epidemics, minor influenza genetic changes occur (antigenic drift). These minor changes are relatively predictable and clinicians are able to develop effective flu vaccines every year. However, highly virulent duel-specie strains are a challenge to quarantine. Due to the sudden and drastic antigenic shifts that are observed in rare/novel hybrid viral strains, making vaccine predictions are no longer possible and pre-designed flu shots are ineffective.
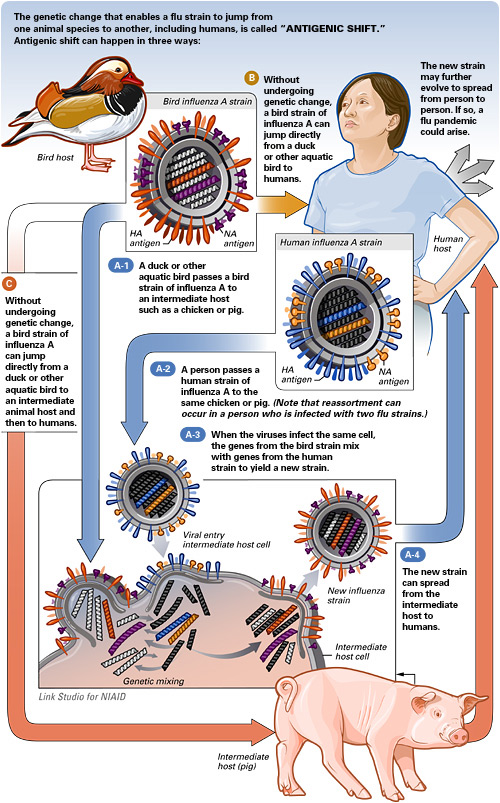
Credit: National Institute of Allergy and Infectious Diseases (NIAID)
Another reason as to why researchers are unable to predict epidemics is that they are unclear as to what key mechanisms are required for viral emergence. The truth is that there really aren’t a lot of observable trends to base predictions off of and there isn't a way to determine how susceptible a potential host is for a given virus.
Evolution, for example, seems as if it would be one of the mechanisms required for cross-species transmission. In order to successfully infect a new host, the virus must be able to effectively replicate in the host’s cells. Such a task is met with multiple barriers including host cell entry via receptor binding which requires genetic compatibility. Overcoming all of these hurdles means there must be a corresponding change in the virion's genetic makeup.
Avian influenza viruses replicate in the gastrointestinal tract while human influenza viruses replicate in the respiratory tract—as conveyed by their differential genetic makeup. What researchers don’t know is if viruses that jump the species barrier are already compatible with the new host or if they evolve compatibility after the fact. Needless to say, evolutionary adaptation to new hosts is a poorly understood process.
Although we know that hybrid influenza viruses are a serious risk, evolutionary changes are not always required for viral emergence in a new host. In the case of the 1918 human influenza epidemic, it is believed that the virus contained eight avian RNA strands. It was also just determined that Influenza A H1N1 is a combination of two pig Influenza viruses. Therefore, cross-species transmissions do not always require antigenic shifts in order to adapt to their new host.
Intuition seemingly breaks down when studying cross-species transmission. It’s logical to assume that the more closely related to a species we are, the more likely it is we will contract viruses from them. Indeed the majority of viruses that humans contract are mammalian in origin, but we have also contracted viruses from birds. HIV is an example of a virus contracted from a closely related species, however, even though we’re most closely related to chimpanzees, our exposure to them is limited. On a global scale, humans are more exposed to rodents which are responsible for a large number of emerged viruses in humans.
Another factor to take into consideration is that more closely related species share the same/similar alleles and other innate immune antibodies, meaning some organisms have pre-immunity to closely related organism viruses. This is in contrast to more distantly related species where less immune system components are similar.
Overall, phylogenetic relationships may not be a crucial factor in cross-species transmission but rather, the occurrence of residing in close proximity to other animals. Also, domesticated animals sometimes act as transitionary viral carriers. SARS emergence for example, originated in bats, but humans contracted the virus from civet cats.
Not knowing factors that determine whether or not a virus will result in a pandemic is a major problem in public health. We realize that it is a very real possibility for a new and rapidly spreading virus to potentially reach numbers to where quarantine is no longer a viable strategy—hence the root cause of mass hysteria. Clearly we must take advised precautions in such times, but epidemics are and have always been inherently part of the human condition and is sometimes out of our control, which if anything, accepting reality can have a calming effect.
References:
Holmes, E. C., Drummond, A. J. (2007). The evolutionary genetics of emergence. CTMI 315:51-66.
Parrish, C. R., Holmes, E. C., Morens, D. M., et al. (2008). Cross-species virus transmission and the emergence of new epidemic diseases. Microlobiology and Molecular Biology Reviews 72:457-470.
Slonczewski, J. L., Foster, J. W., Gillen, K. M. Microbiology: An evolving science. (2009). W. W. Norton&Company, Inc. New York, NY.





Comments